Automated Nucleic Acid Extraction from Stool Samples: A Comprehensive Guide for Researchers and Developers
Automated nucleic acid extraction from stool samples is a critical yet challenging step in molecular diagnostics and microbiome research.
Automated Nucleic Acid Extraction from Stool Samples: A Comprehensive Guide for Researchers and Developers
Abstract
Automated nucleic acid extraction from stool samples is a critical yet challenging step in molecular diagnostics and microbiome research. This article provides a comprehensive overview for researchers and drug development professionals, covering the foundational principles of stool as a complex sample matrix, current automated methodologies including magnetic bead-based systems and novel nanoparticle technologies, strategies for troubleshooting common inhibitors and optimizing protocols, and frameworks for analytical and clinical validation. By synthesizing the latest technological advancements and practical implementation insights, this guide aims to support the development of robust, high-yield extraction workflows for applications ranging from infectious disease diagnostics to microbiome profiling and colorectal cancer screening.
Why Stool is Challenging: Understanding the Complex Matrix for Nucleic Acid Extraction
The automated extraction of nucleic acids from stool samples is a cornerstone of modern molecular diagnostics and microbiome research. However, the complex and inhibitor-rich nature of stool presents significant challenges for consistent and reliable results. A comprehensive understanding of the key interfering substances and the development of robust protocols to neutralize them are critical for the success of downstream applications, such as PCR, sequencing, and the detection of pathogens or host biomarkers. This Application Note details the primary inhibitors found in human stool, provides validated protocols for their mitigation, and presents quantitative data to guide researchers in optimizing automated nucleic acid extraction workflows. The information is framed within the context of advancing research for large-scale screening studies and diagnostic test development [1] [2].
Key Inhibitors and Interfering Substances in Stool
The efficacy of nucleic acid extraction and subsequent molecular analyses is frequently compromised by a variety of substances endogenous to stool. These compounds can co-purify with nucleic acids and inhibit enzymatic reactions. The table below summarizes the major categories of inhibitors, their impact on downstream processes, and their common sources.
Table 1: Key Inhibitors and Interfering Substances in Stool Samples
| Inhibitor Category | Specific Examples | Primary Impact on Downstream Assays | Origin in Stool |
|---|---|---|---|
| Complex Polysaccharides | Hemicelluloses, Pectins | Bind to nucleic acids and enzymes; increase viscosity [3]. | Plant-derived dietary fiber. |
| Bile Salts | Bilirubin, various bile acids | Disrupt enzyme function by denaturing proteins [3]. | Digestive secretions from the liver. |
| Bacterial Metabolites | Short-chain fatty acids, organic acids | Alter pH and disrupt enzymatic activity [3]. | Fermentation by gut microbiota. |
| Pigments | Heme, Bilin | Interfere with spectrophotometric quantification and PCR [3]. | Breakdown of blood cells and chlorophyll. |
| Proteases | Various host and microbial enzymes | Degrade polymerase and other enzymes used in molecular assays [3]. | Pancreatic secretions and gut bacteria. |
| Ionic Substances | Calcium, inorganic salts | Can affect enzymatic efficiency and nucleic acid stability [3]. | Diet, secretions, and microbial activity. |
Quantitative Impact of Inhibitors on DNA Yield and Quality
The presence of inhibitors not only affects amplification but also the initial recovery of nucleic acids. Variability in stool composition leads to significant differences in DNA yield and quality. The following table compiles data from studies that have quantified these parameters, highlighting the challenge of standardization.
Table 2: DNA Yield and Quality from Stool Samples
| Study Reference | Sample Input | Extraction Method | Average DNA Yield (μg/g of stool) | Human DNA Content (%) | Key Challenge Reported |
|---|---|---|---|---|---|
| Protocol (2000) [1] [4] | 2 grams | Phenol/Chloroform Lysis | 9 - 1686 (Highly Variable) | 0.06% - 46% | High variability in total and human DNA yield; presence of PCR inhibitors. |
| Commercial Kit (Patent) [3] | 0.1 - 0.2 grams | Silica-Based Binding with Inhibitor Removal | Not Specified | Not Specified | Neutralization of complex polysaccharides, bilirubin, and bile salts. |
Experimental Protocols for Inhibitor Removal and NA Extraction
Phenol-Chloroform Based DNA Isolation for Large-Scale Studies
This robust protocol is designed for processing large numbers of samples and is independent of the stool collection method, making it suitable for biobanking and population-scale screening [1] [4].
Materials:
- Lysis Buffer: 500 mmol/L Tris-HCl, 100 mmol/L EDTA, 500 mmol/L NaCl [3].
- Lysis Buffer Additives: 2% (w/v) Polyvinylpyrrolidone (PVP) [3], 4 mol/L Guandinium Thiocyanate [3].
- Phenol-Chloroform-Isoamyl Alcohol (25:24:1).
- Glass Beads (100 μm diameter) for mechanical disruption.
- 3 mol/L Sodium Acetate (pH 5.2).
- 100% and 70% Ethanol.
- Nuclease-Free Water.
Procedure:
- Homogenization: Weigh 2 grams of "as dry as possible" stool and suspend in 10 mL of Lysis Buffer. Vortex vigorously for 5 minutes.
- Mechanical Lysis: Add 1 g of glass beads to the suspension and agitate on a mechanical bead-beater for 3 minutes to disrupt tough cell walls and stool matrix.
- Centrifugation: Centrifuge the lysate at 5,000 x g for 10 minutes at room temperature. Transfer the supernatant to a fresh tube.
- Organic Extraction: Add an equal volume of Phenol-Chloroform-Isoamyl Alcohol to the supernatant. Mix thoroughly by vortexing for 2 minutes. Centrifuge at 12,000 x g for 15 minutes at 4°C.
- DNA Precipitation: Carefully transfer the upper aqueous phase to a new tube. Add 0.1 volumes of 3 mol/L Sodium Acetate and 2 volumes of 100% ethanol. Mix and incubate at -20°C for 1 hour.
- DNA Washing: Centrifuge at 12,000 x g for 15 minutes at 4°C to pellet the DNA. Carefully decant the supernatant and wash the pellet with 5 mL of 70% ethanol. Centrifuge again for 5 minutes and carefully pour off the ethanol.
- DNA Resuspension: Air-dry the pellet for 10-15 minutes and resuspend in 100 μL of nuclease-free water. Quantify DNA using a fluorometric method.
Silica-Binding Protocol with Integrated Inhibitor Neutralization
This method is optimized for automated nucleic acid extraction systems and focuses on purifying DNA while retaining compatibility with sensitive downstream applications like real-time PCR [3].
Materials:
- Inhibitor Removal Buffer: 50 mmol/L Tris-HCl, 25 mmol/L EDTA, 4.2 mol/L Guandinium Thiocyanate, 2% (w/v) PVP, 0.5% (v/v) Triton X-100 [3].
- Binding Buffer: 50 mmol/L Tris-HCl, 4.2 mol/L Guandinium Thiocyanate, 30% (v/v) Isopropanol [3].
- Wash Buffer 1: 50 mmol/L Tris-HCl, 5.2 mol/L Guandinium Thiocyanate, 60% (v/v) Ethanol [3].
- Wash Buffer 2: 10 mmol/L Tris-HCl, 80% (v/v) Ethanol, 100 mmol/L NaCl [3].
- Silica-Membrane Binding Columns.
- Nuclease-Free Water.
Procedure:
- Lysis and Inhibitor Binding: Homogenize 0.1-0.2 grams of stool in 1 mL of Inhibitor Removal Buffer. Vortex for 2 minutes. Incubate at 70°C for 5 minutes to enhance inhibitor dissociation.
- Clarification: Centrifuge the lysate at 12,000 x g for 5 minutes to pellet insoluble debris and inhibitor complexes. Transfer the clarified supernatant to a new tube.
- DNA Binding: Add 2 volumes of Binding Buffer to the supernatant and mix by pipetting. Load the mixture onto a silica-membrane column and centrifuge at 11,000 x g for 1 minute. Discard the flow-through.
- Washing: Add 500 μL of Wash Buffer 1 to the column. Centrifuge at 11,000 x g for 1 minute. Discard the flow-through. Add 500 μL of Wash Buffer 2 and repeat the centrifugation. Discard the flow-through.
- Final Wash and Elution: Perform an additional centrifugation step with the empty column for 2 minutes to dry the membrane completely. Elute the DNA by adding 50-100 μL of Nuclease-Free Water to the center of the membrane, incubating for 2 minutes, and centrifuging at 11,000 x g for 1 minute.
The Scientist's Toolkit: Essential Research Reagent Solutions
Successful nucleic acid extraction from stool relies on a specific set of reagents designed to lyse cells, protect nucleic acids, and sequester inhibitors.
Table 3: Key Research Reagent Solutions for Stool NA Extraction
| Reagent | Function | Mechanism of Action |
|---|---|---|
| Guandinium Thiocyanate | Chaotropic Salt / Denaturant | Denatures proteins and nucleases, disrupts cell membranes, and inactivates pathogens [3]. |
| Polyvinylpyrrolidone (PVP) | Inhibitor Binding | Binds to polyphenols and pigments (e.g., heme) via hydrogen bonding, preventing their co-purification with DNA [3]. |
| EDTA (Chelating Agent) | Nuclease Inhibitor | Chelates Mg²⁺ ions, which are essential cofactors for many DNases and RNases, thereby protecting nucleic acids from degradation [3]. |
| Silica-Based Membranes | Nucleic Acid Binding | In the presence of high-concentration chaotropic salts, nucleic acids bind to the silica matrix while impurities are washed away [3]. |
| Polyethylene Glycol (PEG) | Precipitation Aid | Acts as a crowding agent to precipitate nucleic acids, often used in larger-scale or metagenomic preparations. |
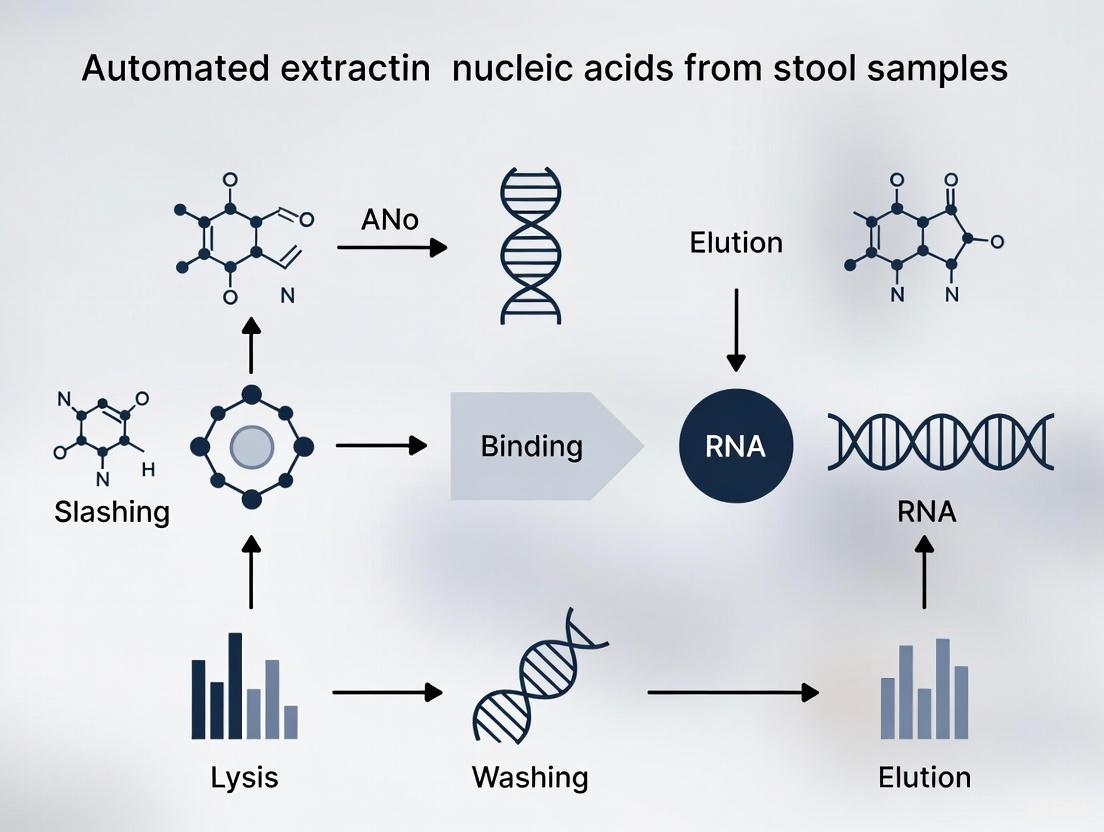
Workflow and Inhibitor Pathways
The following diagrams illustrate the complete workflow for nucleic acid extraction from stool and the specific points at which key inhibitors interfere, alongside the corresponding neutralization strategies.
Stool NA Extraction & Inhibition Management Workflow
Application Notes
Automated nucleic acid extraction from stool samples is a cornerstone technique that provides the foundational genetic material for advancements across infectious disease, microbiome research, and oncology. The transition from manual to automated methods significantly enhances throughput, improves reproducibility, and reduces inter-sample variability, which is critical for generating robust data in both research and clinical diagnostics [5].
Application in Infectious Disease
In infectious disease diagnostics, automated extractors enable sensitive and specific detection of viral, bacterial, and fungal pathogens from complex stool samples. The primary challenge is the efficient removal of PCR inhibitors commonly found in stool to ensure reliable downstream molecular detection.
- Viral Detection: Automated systems have been validated for detecting enteric viruses like norovirus and rotavirus. A comparative study of five platforms demonstrated that while all platforms yielded comparable results for norovirus RNA extraction, the performance could be impaired by inhibitory substances in some samples, highlighting the need for optimized inhibitor removal technologies [6]. In another evaluation focusing on rotavirus RNA, extracts prepared using the MagNA Pure Compact instrument yielded the most consistent results in both qRT-PCR and conventional RT-PCR assays [7].
- Bacterial and Fungal Detection: The integration of bead-beating, a mechanical lysis step, is crucial for the effective rupture of tough microbial cell walls, including those of Gram-positive bacteria and fungal spores. Systems like the KingFisher Apex, which incorporate this step, provide a more comprehensive profile of the microbial community in stool, which is essential for diagnosing bacterial and fungal infections [5].
Application in Microbiome Research
The field of microbiome research relies on the unbiased extraction of total nucleic acids to characterize the diverse communities of bacteria, viruses, and fungi inhabiting the human gut. The choice of extraction method directly influences the observed microbial community structure.
- Impact of Lysis Method: The omission of bead-beating can lead to a significant under-representation of Gram-positive bacteria, such as Firmicutes, in subsequent sequencing data. A 2024 study confirmed that bead-beating provided an incremental yield and more effective lysis of stool samples compared to lysis buffer alone, regardless of the automated extractor used [5].
- Quantitative Microbiome Profiling (QMP): Moving beyond relative abundance data, QMP integrates absolute quantification of microbial abundances into sequencing data. This can be achieved by:
- Flow Cytometry (QMP): Counting intact microbial cells using flow cytometry [8].
- PMA Treatment (QMP-PMA): Using propidium monoazide to pre-treat samples before DNA isolation, thereby profiling only the composition of intact cells and excluding free extracellular DNA [8].
- qPCR (QMP-qPCR): A cost-effective alternative that uses qPCR targeting the 16S rRNA gene to quantify the total bacterial load [8].
- Sequencing and Analysis: High-quality DNA extracted by kits like the MagMAX Microbiome kit is compatible with next-generation sequencing applications, including 16S rRNA gene amplicon sequencing and shotgun metagenomics. This allows researchers to generate species-level profiles to study changes in the microbiome associated with conditions like ulcerative colitis or dietary interventions [9].
Application in Oncology
The gut microbiome is increasingly recognized as a key modulator of cancer development, progression, and response to therapy. Automated nucleic acid extraction from stool enables the study of the microbiome's role in oncogenesis and its potential as a source of biomarkers or therapeutic targets.
- Microbial Drivers of Cancer: An estimated 28.7% of cancers in sub-Saharan Africa are linked to known infectious triggers, including HPV, hepatitis viruses, and H. pylori [10]. Research is now exploring the potential oncogenic roles of other microbes, such as Fusobacterium nucleatum, in various cancers [10].
- The Tumor Microbiome (TM): The TM comprises bacteria, fungi, and viruses within tumor tissues. It can modulate the tumor microenvironment by influencing critical signaling pathways like WNT/β-catenin, NF-κB, and TLRs, thereby affecting cancer progression and response to treatments like immunotherapy [11].
- Mechanisms of Oncogenesis: Microbes can contribute to cancer through multiple mechanisms, including:
Table 1: Comparison of Automated Nucleic Acid Extraction Systems for Stool Samples
| Extractor & Kit | Technology | Throughput (samples/run) | Bead-Beating | Processing Time (for 16 samples) | Key Performance Findings |
|---|---|---|---|---|---|
| KingFisher Apex (MagMAX Microbiome Ultra Kit) | Magnetic Beads | 1–96 | Yes, required | ~40 min | Effective lysis; high-quality DNA for NGS; low inter-sample variability [9] [5] |
| Maxwell RSC 16 (Maxwell RSC Fecal Microbiome DNA Kit) | Magnetic Beads (Pre-packed Cartridges) | 1–16 | Optional | ~42 min | Good yield and purity; performance improved with bead-beating [5] |
| GenePure Pro (MagaBio Fecal Pathogens DNA Purification Kit) | Magnetic Beads (Pre-packed Plate) | 1–32 | Optional | ~35 min | Differences in yield and inter-sample variability observed compared to other systems [5] |
| EZ2 Connect (EZ2 PowerFecal Pro DNA/RNA Kit) | Magnetic Beads (Pre-filled Cartridges) | Up to 24 | Yes, via bead beating | Information Missing | High yields of inhibitor-free DNA/RNA; suitable for PCR and NGS [12] |
Table 2: Quantitative Microbiome Profiling (QMP) Approaches
| Method | Description | Advantages | Limitations |
|---|---|---|---|
| QMP | Combines flow cytometry cell counting with 16S sequencing. | Overcomes compositional bias of relative abundance data. | Does not capture free extracellular DNA, which can introduce bias if its composition differs from intact cells [8]. |
| QMP-PMA | QMP with propidium monoazide pre-treatment to bind to DNA from dead cells. | Profiles only intact, viable cells. | Adds a processing step; may not penetrate all complex samples equally [8]. |
| QMP-qPCR | Uses qPCR to quantify 16S rRNA gene copies for absolute abundance. | Cost-effective, simple, and accessible. | Lower sensitivity; may only detect 2-fold changes in microbial load [8]. |
Experimental Protocols
Protocol: Automated Nucleic Acid Extraction from Stool Using a Magnetic Bead-Based System
This protocol is adapted for systems like the KingFisher Apex with the MagMAX Microbiome Ultra Nucleic Acid Isolation Kit or similar [9] [5].
2.1.1 Research Reagent Solutions
Table 3: Essential Materials for Automated Stool DNA Extraction
| Item | Function | Example Product |
|---|---|---|
| Automated Nucleic Acid Extractor | Performs automated binding, washing, and elution of nucleic acids. | KingFisher Apex, GenePure Pro, Maxwell RSC 16 [5] |
| Extraction Kit | Provides optimized buffers, magnetic beads, and reagents. | MagMAX Microbiome Ultra Kit, Maxwell RSC Fecal Microbiome Kit [9] [5] |
| Lysis Buffer with Inhibitor Removal | Disrupts microbial and stool cells, inactivates nucleases, and begins removal of PCR inhibitors. | Lysis buffer from commercial kits [12] |
| Proteinase K | Digests proteins and degrades nucleases. | Supplied in many kits or purchased separately [5] |
| Magnetic Silica Beads | Bind nucleic acids in the presence of chaotropic salts for purification. | Supplied in magnetic bead-based kits [9] [12] |
| Wash Buffers | Remove salts, proteins, and other impurities from the bound nucleic acids. | Supplied in kits [9] [5] |
| Nuclease-Free Elution Buffer | Releases purified nucleic acids from the magnetic beads into a stable solution. | Low-salt buffer like Tris-EDTA (TE) or kit-supplied elution buffer [9] [5] |
| Bead-Beater/Homogenizer | Mechanically lyses tough microbial cell walls using beads. | Omni Bead Ruptor 96, BeadBug, FastPrep-24 [9] [5] |
| Sample Preservation Reagent | Stabilizes nucleic acids in stool at room temperature during storage/transport. | DNA/RNA Shield [5] |
2.1.2 Procedure
Sample Preparation:
Mechanical Lysis (Bead-Beating):
- Transfer the sample to a tube or plate suitable for bead-beating.
- Perform mechanical homogenization. Example settings from the MagMAX protocol are provided in Table 4 [9].
- Centrifuge the lysate briefly to pellet debris.
Lysate Transfer:
- Transfer a defined volume of supernatant (e.g., 300 µL) to a deep-well plate or the specific plate/cartridge required by the automated extractor [5].
Automated Extraction:
- Load the plate/cartridge onto the instrument along with the necessary reagents (e.g., magnetic beads, wash buffers, elution buffer).
- Select and run the appropriate manufacturer-provided protocol for DNA or total nucleic acid extraction. A typical workflow is illustrated in Diagram 2.
Post-Elution:
- Retrieve the eluted nucleic acids (typically in 50-100 µL elution buffer).
- Quantify the DNA concentration and assess purity using a fluorometer (e.g., Qubit) and spectrophotometer (e.g., NanoDrop), respectively [5].
- Store purified DNA at -80°C until downstream application.
Table 4: Example Bead-Beating Settings for Microbial Lysis [9]
| Instrument | Setting |
|---|---|
| Omni Bead Ruptor 96 | 30 Hz for 2 minutes |
| Mini Bead Beater 96 | 2 minutes |
| Bead Bug | 4 minutes at 4 m/s |
| Vortex with plate adapter | 5 minutes at 2,000 rpm |
Protocol: 16S rRNA Gene Amplicon Sequencing for Microbiome Analysis
This protocol follows the methodology used in the 2024 comparative study [5].
2.2.1 Library Preparation:
- Amplification: Amplify the hypervariable V3-V4 region of the 16S rRNA gene from the extracted DNA using primers 338F (5'-CCTACGGRRBGCASCAGKVRVGAAT-3') and 806R (5'-GGACTACNVGGGTWTCTAATCC-3').
- PCR Conditions:
- Initial Denaturation: 95°C for 3 min.
- 28 Cycles of:
- Denaturation: 95°C for 30 s.
- Annealing: 55°C for 30 s.
- Elongation: 72°C for 30 s.
- Final Elongation: 72°C for 5 min [5].
- Library Construction: Use a library preparation kit (e.g., Nextera DNA Library Prep Kit) to index the amplicons.
- Sequencing: Pool the indexed libraries and perform sequencing on a platform such as the Illumina MiSeq (2 × 300 bp paired-end).
2.2.2 Bioinformatic Analysis:
- Process raw sequencing reads using a pipeline (e.g., QIIME 2, mothur) to perform quality filtering, denoising, and chimera removal.
- Cluster sequences into Amplicon Sequence Variants (ASVs) or Operational Taxonomic Units (OTUs).
- Assign taxonomy using a reference database (e.g., SILVA, Greengenes).
- Perform diversity analysis (alpha and beta-diversity) and differential abundance testing to compare microbial communities between sample groups.
Pathway and Workflow Visualizations
Automated nucleic acid extraction from stool samples presents a significant challenge in molecular diagnostics and research. Stool is a complex mixture containing a wide range of PCR inhibitors, including bile pigments, complex carbohydrates, and bacterial metabolites [6] [13]. The effectiveness of nucleic acid extraction directly impacts the sensitivity and reliability of downstream applications such as PCR, microarray analysis, and next-generation sequencing [14]. Within the context of a broader thesis on automated extraction, this application note delineates the essential requirements for effective nucleic acid isolation, focusing on the critical triumvirate of purity, yield, and inhibitor removal. Successful extraction must deliver nucleic acids free from contaminants that interfere with enzymatic reactions, provide sufficient yield for intended applications, and effectively remove substances that inhibit amplification [14].
Key Requirements for Effective Extraction
The table below summarizes the core parameters and their significance in automated nucleic acid extraction from stool samples.
Table 1: Essential Requirements for Effective Nucleic Acid Extraction from Stool Samples
| Requirement | Description | Impact on Downstream Applications |
|---|---|---|
| Purity | Absence of contaminants (e.g., proteins, salts, organic compounds) that absorb at 260 nm or inhibit enzymes. | Essential for accurate spectrophotometric quantification and robust performance in PCR and sequencing [15]. |
| Yield | The total amount of nucleic acid recovered from the starting sample. | Critical for detecting low-abundance targets and enabling multiple downstream analyses from a single extraction [16]. |
| Inhibitor Removal | Effective elimination of stool-specific inhibitors such as bile salts, complex polysaccharides, and heme. | Paramount for achieving high sensitivity in molecular diagnostics, as inhibitors can cause false-negative results [6] [17]. |
| Integrity | Preservation of nucleic acid fragment length and structure. | Important for long-range PCR and sequencing applications that require high-molecular-weight DNA [15]. |
| Automation Compatibility | Suitability for integration into automated, high-throughput workflows. | Reduces hands-on time, minimizes human error, and increases reproducibility in research and clinical settings [18]. |
Experimental Protocols for Validation
Protocol: Evaluating Extraction Efficiency and Inhibitor Removal
This protocol is adapted from methodologies used to validate and optimize automated extraction systems for complex samples [6] [17].
1. Sample Preparation:
- Obtain stool samples from healthy donors and homogenize in a suitable buffer (e.g., PBS or commercial stool transport medium).
- Spike samples with a known quantity of an exogenous control (e.g., a non-human pathogen or synthetic nucleic acid) prior to extraction to monitor recovery efficiency.
- Aliquot samples for parallel extraction across different platforms or conditions.
2. Automated Nucleic Acid Extraction:
- Platform Examples: Systems such as NucliSENS easyMAG (bioMerieux), MagNA Pure (Roche), or QiaSymphony (Qiagen) can be employed [6].
- Lysis: Use a lysis buffer containing a strong chaotropic agent like guanidinium thiocyanate. For robust lysis of hardy organisms (e.g., spores, fungi), incorporate a mechanical disruption step using a bead beater (e.g., 2-5 minutes at high speed) prior to automation [17] [19].
- Extraction: Follow manufacturer-recommended protocols. A key parameter to optimize is the input volume of magnetic silica beads; for high-density samples, 140 µL may be superior to 50 µL for maximizing yield [17].
- Elution: Elute nucleic acids in a low-salt buffer (e.g., TE buffer or nuclease-free water) at a pH >8.0, using a small volume (e.g., 50-100 µL) to maximize concentration [17].
3. Downstream Quantification and Qualification:
- Spectrophotometry: Measure A260/A280 and A260/A230 ratios to assess purity. Ideal ratios are ~1.8 and >2.0, respectively [15].
- Fluorometry: Use dye-based quantification (e.g., Qubit) for a more accurate measurement of nucleic acid concentration, as it is less affected by contaminants.
- qPCR: Perform quantitative PCR targeting a ubiquitous bacterial gene (e.g., 16S rRNA) and the spiked exogenous control. The cycle threshold (Ct) values allow for the relative quantification of yield and direct assessment of PCR inhibition. A significant delay in the Ct of the spiked control in sample extracts versus a clean control indicates the presence of residual inhibitors [6] [17].
Workflow: Automated NA Extraction from Stool
The following diagram illustrates the optimized workflow for automated nucleic acid extraction from stool samples, integrating key steps for ensuring purity, yield, and effective inhibitor removal.
The Scientist's Toolkit: Research Reagent Solutions
The following table details key reagents and materials essential for successful automated nucleic acid extraction from challenging stool samples.
Table 2: Essential Research Reagents and Materials for Automated NA Extraction
| Item | Function | Application Note |
|---|---|---|
| Magnetic Silica Beads | Solid-phase matrix that binds nucleic acids in the presence of chaotropic salts. | The surface area and concentration of beads are critical; higher bead volumes (e.g., 140 µL) improve yield from inhibitor-rich samples [17]. |
| Chaotropic Lysis Buffer | Disrupts cells, inactivates nucleases, and enables nucleic acid binding to silica. | Guanidinium thiocyanate-based buffers are highly effective for denaturing proteins and inactivating pathogens in stool samples [16] [17]. |
| Inhibitor Removal Buffers | Wash buffers designed to remove specific classes of contaminants. | A wash with a buffer containing guanidinium maintains binding stringency, while a final high-salt ethanol wash removes residual salts and other impurities [17]. |
| Bead-Beating Tubes | Tubes containing ceramic or glass beads for mechanical disruption. | Essential for breaking down hardy structures like fungal cell walls, protozoan cysts, and bacterial spores in stool prior to automated extraction [17] [19]. |
| External Positive Control | A known quantity of non-target nucleic acid spiked into the sample. | Used to monitor extraction efficiency and detect the presence of PCR inhibitors by comparing Cq values to a clean extraction [6] [17]. |
Performance Data and Comparison
Comparative studies of automated extraction platforms provide critical insights into their performance with stool samples. The following table summarizes quantitative data from a study comparing five automated systems for the extraction of viral RNA from stool.
Table 3: Comparison of Automated Extraction Platforms for Norovirus RNA from Stool [6]
| Extraction Platform | Total Samples (n=39) | Samples Positive on All Platforms | Notes on Inhibition |
|---|---|---|---|
| easyMAG (bioMerieux) | 39 | 36 | Some samples showed inhibition. |
| m2000sp (Abbott) | 39 | 36 | Some samples showed inhibition. |
| MagNA Pure LC 2.0 (Roche) | 39 | 36 | Some samples showed inhibition. |
| QiaSymphony (Qiagen) | 39 | 36 | Some samples showed inhibition. |
| VERSANT kPCR (Siemens) | 39 | 39 | All samples tested positive for both target and internal control. |
Optimization of established methods can lead to significant improvements. Recent research on a magnetic silica bead-based method (SHIFT-SP) demonstrated that adjusting the binding buffer to a lower pH (4.1 versus 8.6) dramatically increased DNA binding efficiency from 84.3% to 98.2% [16]. Furthermore, replacing orbital shaking with a rapid "tip-based" mixing method reduced the binding time from 5 minutes to 1 minute while achieving superior yield, highlighting the impact of optimizing physical parameters in addition to chemical ones [16]. For stool samples, the MagMAX Microbiome kit, which employs bead-beating lysis and magnetic bead-based purification, has been shown to effectively isolate high-quality total nucleic acids compatible with downstream qPCR and NGS applications, as evidenced by clear electrophoretic bands and high RIN scores [19].
The analysis of nucleic acids from stool samples is a cornerstone of modern molecular research, enabling advancements in areas ranging from human microbiome studies to non-invasive cancer detection. Stool represents one of the most complex sample matrices, containing a diverse mixture of microorganisms, host cells, dietary residues, and potent PCR inhibitors. The evolution of nucleic acid extraction technologies from manual protocols to sophisticated automated systems has been driven by the need for higher throughput, improved reproducibility, and more robust results in the face of these challenges. This evolution is particularly critical for large-scale studies and clinical applications where batch effects and inter-sample variation can confound biological interpretations [5] [20]. The transition to automation represents not merely a convenience but a fundamental methodological shift that enhances data quality and reliability while accommodating the increasing scale of biomedical research.
The Technological Shift: From Manual to Automated Extraction
Limitations of Manual Extraction Methods
Traditional manual extraction methods, often based on silica spin columns or phenol-chloroform phase separation, present significant limitations for high-throughput stool analysis. These methods are labor-intensive, time-consuming, and susceptible to human error and inter-operator variability. The FastDNA Spin Kit for Soil, a commonly used manual method for challenging samples like stool, requires approximately 100 minutes of hands-on processing time for just 16 samples and involves multiple centrifugation, incubation, and transfer steps that introduce opportunities for contamination and inconsistency [5] [20]. Additionally, manual methods struggle with the efficient lysis of diverse microbial communities in stool, particularly robust Gram-positive bacteria and bacterial spores, without incorporating dedicated mechanical disruption steps [20].
Advantages of Automated Magnetic Bead-Based Systems
Automated nucleic acid extraction systems have primarily standardized around magnetic bead-based technology, which offers several fundamental advantages for stool analysis:
- Higher Purity and Yields: Magnetic beads provide exceptional binding capacity with thorough exposure to target molecules during mixing and washing steps, allowing for efficient capture and release of nucleic acids [21].
- Enhanced Reproducibility: Standardized protocols involving fewer steps enable consistent and reproducible results with reduced processing time and minimal technical variation [21] [5].
- Scalability and Flexibility: Automation compatibility facilitates high-throughput processing, with systems capable of handling from 16 to 384 samples per run [21] [22].
- Gentle Separation: Without columns, filters, and excessive centrifugation, the technology minimizes potential clogging and helps prevent shearing of sensitive biomolecules [21].
- Superior Inhibitor Removal: Specialized chemistries and wash buffers effectively remove PCR inhibitors common in stool samples, such as bile salts and complex polysaccharides [12] [23].
The Critical Role of Bead-Beating in Stool Analysis
A key consideration in automating stool nucleic acid extraction is the integration of effective mechanical lysis. Research demonstrates that bead-beating provides incremental yield by effectively lysing a broader representation of microbial cells in stool samples compared to chemical lysis buffer alone [5] [20]. Differential abundance analysis reveals a greater representation of Gram-positive bacteria in samples subjected to mechanical lysis, regardless of the automated extraction system used [20]. This finding is significant because inadequate lysis of Gram-positive bacteria can introduce substantial bias in microbiome studies, potentially missing important biological signatures. While not all automated systems incorporate onboard bead-beating, many protocols now include this as a essential pre-processing step to ensure comprehensive lysis of the diverse microbial communities present in stool [20].
Comparative Performance of Automated Extraction Systems
System Comparisons and Technical Specifications
Recent methodological comparisons provide valuable insights into the performance characteristics of different automated platforms for stool analysis. A 2024 systematic evaluation compared three commercial nucleic acid extractors—Bioer GenePure Pro, Promega Maxwell RSC 16, and Thermo Fisher KingFisher Apex—using both human fecal samples and mock microbial communities [5] [20]. The study assessed DNA yield, DNA purity, and 16S rRNA gene amplicon sequencing results, revealing important differences in system performance and downstream applications.
Table 1: Comparison of Automated Nucleic Acid Extraction Systems for Stool Samples
| Extraction System | Throughput (samples/run) | Processing Time (for 16 samples) | Bead-Beating Compatibility | Technology Platform | Key Applications Demonstrated |
|---|---|---|---|---|---|
| Bioer GenePure Pro | 1-32 | ~35 minutes | External (required) | Magnetic bead-based | Microbiome analysis, pathogen detection [5] |
| Promega Maxwell RSC 16 | 1-16 | ~42 minutes | External (required) | Magnetic bead-based | Microbiome analysis, sequential DNA/RNA isolation [5] [21] |
| Thermo Fisher KingFisher Apex | 1-96 | ~40 minutes | Integrated or external | Magnetic bead-based | High-throughput microbiome, virome studies [5] [21] |
| QIAGEN EZ2 Connect | 1-24 | Protocol-dependent | Integrated (Tissuelyser) | Magnetic particle technology | Microbial DNA/RNA, total nucleic acids [12] |
| MGI MGISP-NE384 | 96-384 | High-throughput | External | Magnetic rod technology | Large-scale epidemiology, biobanking [22] |
Quantitative Performance Metrics
The performance evaluation of automated systems reveals critical differences in extraction efficiency, purity, and downstream analytical outcomes. When comparing DNA yield across systems, the EZ2 PowerFecal Pro DNA/RNA Kit demonstrated the highest DNA yields across four different stool samples compared to alternative suppliers [12]. In the three-system comparison, all automated extractors showed differences in terms of yield, inter-sample variability, and subsequent sequencing readouts [20]. These technical differences translated into meaningful variations in 16S rRNA gene amplicon results, highlighting the importance of selecting appropriate extraction methods for specific research applications.
Table 2: Performance Metrics of Automated Extraction Systems for Stool Analysis
| Performance Metric | KingFisher Apex | Maxwell RSC 16 | GenePure Pro | Manual Column-Based |
|---|---|---|---|---|
| Average DNA Yield | Variable by kit | Variable by kit | Variable by kit | Baseline reference [20] |
| Inter-Sample Variability | Lower | Lower | Lower | Higher [5] [20] |
| Process Contamination | Reduced (closed system) | Reduced (closed system) | Reduced (closed system) | More frequent [5] |
| Gram-positive Bacteria Representation | Higher with bead-beating | Higher with bead-beating | Higher with bead-beating | Dependent on protocol [20] |
| Downstream Sequencing Quality | High with appropriate kit | High with appropriate kit | High with appropriate kit | Variable [20] |
| Sample Preparation Time (16 samples) | ~40 minutes | ~35 minutes | ~25 minutes | ~100 minutes [5] |
Specialized Applications and Methodological Adaptations
Different research applications require specialized approaches to nucleic acid extraction from stool:
Virome Studies: Research on the DNA virome from fecal samples often employs specialized protocols for virus-like particle (VLP) enrichment prior to nucleic acid extraction. This typically involves filtration through 0.45 µm and 0.22 µm filters, treatment with DNase to remove free DNA, and subsequent phenol-chloroform extraction [24]. Such specialized procedures demonstrate that even with automated platforms, certain applications require tailored pre-processing steps.
RNA Extraction for Cancer Biomarkers: The detection of colorectal cancer-associated immune genes in stool requires optimized RNA extraction protocols. A 2024 study found that a combination of the Stool total RNA purification kit (Norgen) with the Superscript III one-step RT-PCR kit (Invitrogen) provided high RNA purity and sensitive mRNA detection [25]. The low abundance of human mRNA in stool, relative to microbial RNA, necessitates very sensitive approaches with high specificity to avoid cross-reactivity.
Inhibitor Removal Technologies: Advanced materials are being developed to address the challenge of PCR inhibitors in stool. A 2025 study demonstrated that Fe-doped mesoporous silica nanoparticle (Fe-MSN) columns provided more efficient extraction than conventional methods, yielding higher RNA purity and lower Ct values in RT-PCR [23]. This emerging technology showed a 5-fold decrease in the Ct value of different studied genes compared to a commercial extraction kit.
Detailed Experimental Protocols
Automated DNA Extraction for Microbiome Analysis
Protocol Title: Automated DNA Extraction from Human Stool Samples for 16S rRNA Gene Sequencing
Principle: This protocol utilizes magnetic bead-based technology to isolate high-quality genomic DNA from human stool samples, suitable for downstream applications including qPCR and next-generation sequencing. The method incorporates mechanical bead-beating for comprehensive lysis of diverse microbial populations [5] [20].
Materials and Reagents:
- Stool samples preserved in DNA/RNA Shield or similar preservation reagent
- MagMAX Microbiome Ultra Kit (Thermo Fisher Scientific) or equivalent
- KingFisher Apex System (Thermo Fisher Scientific) or compatible automated extractor
- FastPrep-24 5G Bead Beating Grinder (MP-Biomedicals) or equivalent
- DNA/RNA Shield Fecal Collection Tubes (Zymo Research Corp)
- Nuclease-free water
- Ethanol (96-100%)
- Microcentrifuge tubes (1.5-2 mL)
Procedure:
- Sample Preparation: Thaw frozen fecal samples at room temperature. For each sample, aliquot 300 µL of fecal DNA shield mixture into a lysing matrix tube containing silica beads.
- Mechanical Lysis: Homogenize samples using the FastPrep-24 system at 6.0 m/s for 40 seconds to ensure comprehensive disruption of microbial cells, including difficult-to-lyse Gram-positive bacteria.
- Centrifugation: Centrifuge the lysate at 14,000 × g for 5 minutes to pellet debris while leaving nucleic acids in the supernatant.
- Automated Extraction Setup: Transfer 200 µL of supernatant to a deep-well plate compatible with the automated extraction system. Add recommended volumes of lysis/binding solution, magnetic beads, and wash buffers according to kit specifications.
- Automated Extraction: Run the appropriate program on the automated extractor (e.g., KingFisher Apex). Typical programs include:
- Binding incubation: 10-15 minutes with mixing
- Two wash steps with wash buffers
- Drying step: 5 minutes
- Elution: 5 minutes in nuclease-free water or low-EDTA TE buffer
- DNA Quantification and Quality Assessment: Measure DNA concentration using fluorometric methods (e.g., Qubit dsDNA HS assay) and purity using spectrophotometry (e.g., NanoDrop). Store purified DNA at -80°C until library preparation.
Troubleshooting Notes:
- Low DNA yield may indicate insufficient bead-beating or incomplete sample homogenization
- High inhibitor carryover may require additional wash steps or dilution of extracts
- For difficult samples, increasing the bead-beating time or using smaller bead sizes may improve lysis efficiency
Automated RNA Extraction for Gene Expression Analysis
Protocol Title: Automated RNA Extraction from Stool for mRNA Biomarker Detection
Principle: This protocol isolates high-quality total RNA from stool samples for detection of human mRNA transcripts, enabling non-invasive monitoring of colorectal cancer-associated immune genes. The method combines effective inhibitor removal with preservation of labile RNA molecules [25].
Materials and Reagents:
- Stool samples preserved in RNAlater
- EZ2 PowerFecal Pro DNA/RNA Kit (QIAGEN) or equivalent
- EZ2 Connect Instrument (QIAGEN)
- RNase-free DNase Set (for DNA-only removal)
- β-mercaptoethanol
- Ethanol (96-100%)
- Nuclease-free water
Procedure:
- Sample Homogenization: Weigh 50-100 mg of stool and transfer to a tube containing lysis buffer and β-mercaptoethanol. Homogenize thoroughly using a vortex mixer with beads or a commercial homogenizer.
- Inhibitor Removal: Add the proprietary inhibitor removal solution and mix thoroughly. Centrifuge to pellet inhibitors and transfer the supernatant to a new tube.
- Automated Extraction Setup: Load the supernatant onto the EZ2 Connect cartridge along with the appropriate reagents for RNA isolation. For DNA-free RNA, ensure DNase is included in the protocol.
- Automated Extraction: Run the RNA isolation protocol on the EZ2 Connect system. The automated process includes:
- Nucleic acid binding to magnetic particles
- Optional DNase digestion (if selecting for RNA only)
- Multiple wash steps to remove contaminants
- Elution in RNase-free water
- RNA Quality Assessment: Measure RNA concentration using fluorometry (e.g., Qubit RNA HS assay) and purity using spectrophotometry. Assess RNA integrity if necessary using automated electrophoresis systems.
- Downstream Application: Use purified RNA immediately in RT-PCR reactions or store at -80°C for future use.
Application Notes:
- This protocol has been successfully used to detect immune genes (CXCL1, IL8, IL1B, IL6, PTGS2, and SPP1) in colorectal cancer research [25]
- For optimal results, process samples immediately after collection or ensure proper preservation in RNAlater
- Include appropriate negative controls to monitor for contamination during extraction
Workflow Visualization: Automated Nucleic Acid Extraction from Stool
Automated Stool Nucleic Acid Extraction Workflow
This workflow diagram illustrates the integrated process of automated nucleic acid extraction from stool samples, highlighting the manual pre-processing requirements, the fully automated extraction steps, and the essential quality control procedures necessary for successful downstream applications.
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Essential Reagents and Kits for Automated Nucleic Acid Extraction from Stool
| Product Name | Manufacturer | Primary Application | Key Features | Compatible Systems |
|---|---|---|---|---|
| MagMAX Microbiome Ultra Kit | Thermo Fisher Scientific | DNA extraction from microbiome samples | Optimized for difficult-to-lyse bacteria; inhibitor removal technology | KingFisher systems [21] [20] |
| EZ2 PowerFecal Pro DNA/RNA Kit | QIAGEN | Simultaneous DNA/RNA extraction | Second-generation Inhibitor Removal Technology; pre-filled cartridges | EZ2 Connect [12] |
| Maxwell RSC Fecal Microbiome DNA Kit | Promega | DNA for microbiome analysis | Efficient lysis chemistry; minimal cross-contamination | Maxwell RSC [5] |
| MagaBio Fecal Pathogens DNA Purification Kit | Bioer Technology | Pathogen detection | Comprehensive pathogen lysis; high sensitivity | GenePure Pro [5] |
| Stool total RNA purification kit | Norgen Biotech | RNA for gene expression | Effective eukaryotic RNA recovery; DNase treatment | Multiple systems [25] |
| NucliSENS EasyMAG system | BioMérieux | RNA for viral detection | Generic protocol for stool; magnetic silica | EasyMAG [25] |
The evolution from manual to automated nucleic acid extraction technologies has fundamentally transformed stool-based molecular research, enabling the high-throughput, reproducible analyses required for modern biomedical science. Automated magnetic bead-based systems address the specific challenges posed by complex stool matrices while providing the scalability needed for large-scale studies. The integration of mechanical bead-beating, advanced inhibitor removal technologies, and standardized protocols has significantly improved the representation of diverse microbial communities and the detection of host biomarkers in stool. As research continues to advance, further innovations in automation, miniaturization, and materials science will continue to enhance our ability to extract meaningful biological information from this challenging but invaluable sample type. The methodologies and systems detailed in this application note provide researchers with the foundational knowledge needed to select and implement appropriate automated extraction strategies for their specific stool-based research applications.
Current Automated Platforms and Emerging Technologies in Practice
Within research frameworks focusing on the automated extraction of nucleic acids from stool samples, solid-phase extraction utilizing magnetic beads has emerged as the dominant methodology. Stool samples represent one of the most complex and challenging sample types, characterized by the presence of extensive PCR inhibitors such as bile salts and complex carbohydrates [14]. Efficient extraction of high-quality nucleic acid is therefore a critical pre-analytical step for downstream applications including qPCR, next-generation sequencing, and microbiome analysis [21]. Magnetic bead-based technology has been widely adopted in this context due to its advantages in automation compatibility, scalability, and ability to deliver high-purity nucleic acids free from common inhibitors found in stool [14] [21].
Core Principles and Advantages
The fundamental principle of magnetic bead-based nucleic acid extraction involves the use of silica-coated magnetic particles that bind nucleic acids in the presence of chaotropic salts [14]. The process can be broken down into four key stages, which are easily integrated into automated liquid handling platforms:
- Lysis: Chemical and/or mechanical disruption of sample cells and viruses to release nucleic acids.
- Binding: Introduction of magnetic beads in a high-salt buffer, promoting the adsorption of nucleic acids onto the silica surface of the beads.
- Washing: Application of a magnetic field to immobilize the bead-nucleic acid complexes while contaminants are removed with wash buffers. This step is crucial for removing PCR inhibitors from complex samples like stool.
- Elution: Resuspension of the purified beads in a low-ionic-strength buffer or water, causing the nucleic acids to dissociate and enter the solution, resulting in a pure eluate [14] [21].
The transition to this method from older techniques is driven by its distinct advantages, particularly for challenging samples and high-throughput environments.
Table 1: Comparison of Nucleic Acid Extraction Methods
| Feature | Phenol-Chloroform | Column-Based Silica | Magnetic Beads (Automated) |
|---|---|---|---|
| Principle | Liquid-phase separation, alcohol precipitation [14] | Solid-phase on silica membrane [14] | Solid-phase on silica-coated magnetic particles [14] [21] |
| Automation Potential | Low | Moderate | High |
| Throughput | Low | Low to Moderate | High to Very High |
| Risk of Contamination | High | Moderate | Low |
| Hands-on Time | High | Moderate | Low |
| Suitability for Complex Samples (e.g., Stool) | Poor (inefficient inhibitor removal) | Moderate | Excellent (efficient washing) [21] |
| Shearing Risk | High (due to vigorous mixing) | Moderate (from centrifugation) | Low (gentle mixing) [21] |
Detailed Protocol for Stool Samples
The following protocol is adapted for automated extraction of nucleic acids from stool samples using a magnetic bead-based system, such as the KingFisher system paired with a dedicated kit (e.g., MagMAX Microbiome Kit) [21].
Research Reagent Solutions
Table 2: Essential Materials and Reagents
| Item | Function / Description |
|---|---|
| Automated Purification System | e.g., KingFisher System. Instrument that moves magnetic beads through various solutions for hands-free purification [21]. |
| Magnetic Bead Kit for Microbiome | e.g., MagMAX Microbiome Kit. Provides optimized buffers and magnetic beads for efficient lysis and purification from complex samples [21]. |
| Lysis Buffer (with Beads) | Contains chaotropic salts to promote nucleic acid binding to beads and other reagents to begin breaking down sample matrix [21]. |
| Wash Buffers | Typically two washes: one with a salt-ethanol solution to remove contaminants, and a second with ethanol to remove residual salts [14] [21]. |
| Elution Buffer | Low-ionic-strength solution (e.g., TE buffer or nuclease-free water) that causes nucleic acids to release from the beads into the solution [14]. |
| Proteinase K | Enzyme added to lysis buffer to digest proteins and enhance cell wall breakdown, especially for Gram-positive bacteria in stool [14] [26]. |
| Bead-Beating Tubes | Tubes containing mechanical beads for homogenization. Critical for disrupting tough bacterial and fungal cell walls in stool microbiota. |
Step-by-Step Workflow
Pre-processing (Manual):
- Sample Homogenization: Suspend a small aliquot (~100-200 mg) of stool in an appropriate transport or lysis buffer. Vortex thoroughly.
- Mechanical Lysis: Transfer an aliquot (e.g., 200 µL) of the homogenate to a bead-beating tube. Homogenize using a Precellys homogenizer or similar (e.g., 3 x 15 s at 10,000 rpm with 10 s intervals) to ensure complete disruption of microbial cells [26].
- Clarification: Centrifuge the lysate briefly at low speed (e.g., 1500 × g for 30 s) to pellet large debris, including undigested food particles and toilet paper common in stool samples [26]. The supernatant is used for automated extraction.
Automated Extraction (on KingFisher System):
- Plate Setup: In a deep-well plate, prepare the following in sequence:
- Well 1: Sample supernatant from the pre-processing step.
- Well 2: Binding solution/Bead mix (magnetic beads in chaotropic salt solution).
- Well 3 & 4: Wash Buffer I and Wash Buffer II.
- Well 5: Elution Buffer.
- Run Method: The automated method follows the logic in the workflow diagram below. The instrument uses a magnetic comb to transfer beads through each solution.
- Eluate Collection: The final eluate (in Well 5) contains purified nucleic acids, ready for quantification and downstream analysis.
Automated Workflow Visualization
Experimental Data and Application
The performance of different extraction protocols can be quantitatively assessed by measuring the yield and detection of specific genetic targets. A recent study evaluating extraction protocols for antibiotic resistance genes (ARGs) in complex wastewater samples provides a relevant model for stool analysis [26].
Table 3: Exemplary Experimental Data Comparing Extraction Protocols for ARG Detection
| Extraction Protocol (EP) | Kit / Method | Sample Input Volume | Target ARG | Mean Concentration (Copies/mL) | Detection Consistency |
|---|---|---|---|---|---|
| EP1 | DNeasy Blood and Tissue Kit [26] | 0.2 mL | tetA | 1.20 x 10⁵ | High |
| EP4 | DNeasy Blood and Tissue Kit (with TP removal) [26] | 1.5 mL | tetA | 1.05 x 10⁵ | High |
| EP1 | DNeasy Blood and Tissue Kit [26] | 0.2 mL | ermB | 9.80 x 10⁴ | High |
| EP5 | AllPrep PowerViral DNA/RNA Kit [26] | 0.2 mL | qnrS | 4.50 x 10³ | Moderate |
| EP10 | AllPrep PowerViral DNA/RNA Kit (with Trizol) [26] | 1.5 mL | qnrS | 5.10 x 10³ | High |
Key Findings from Experimental Data:
- Small Volume Sufficiency: Protocols EP1 and EP5 demonstrate that a starting aliquot as low as 0.2 mL can be sufficient for consistent detection of highly abundant targets, a critical consideration for limited or precious stool samples [26].
- Inhibitor Removal: The high detection consistency for targets like tetA and ermB across different protocols based on magnetic beads highlights the method's efficiency in removing PCR inhibitors, even from complex matrices [26].
- Protocol Optimization: The use of specific additives, such as Trizol in EP10, can enhance the lysis of tough pathogens and improve the detection yield of certain targets, indicating that protocols may be tailored to the specific microbial community or target of interest [26].
Solid-phase extraction with magnetic beads provides a robust, scalable, and efficient foundation for automated nucleic acid purification from stool samples. Its dominance in modern research workflows is justified by the superior purity of the output, the significant reduction in hands-on time, and the direct compatibility with high-throughput instrumentation. As molecular techniques like qPCR and metagenomic sequencing continue to advance, the optimized and automated protocols enabled by magnetic bead technology will remain indispensable for generating reliable and reproducible data in microbiome and pathogen detection research.
The extraction of nucleic acids from complex biological samples like stool is a critical preparatory step in molecular diagnostics and genomics research. The presence of potent PCR inhibitors and diverse microbial communities in stool samples makes them one of the most challenging matrices for nucleic acid isolation. In recent years, functionalized magnetic nanoparticles (MNPs) have emerged as a superior substrate for solid-phase nucleic acid extraction, enabling automation, high throughput, and excellent recovery of both DNA and RNA. Among these, iron oxide-based nanoparticles, particularly magnetite (Fe₃O₄), form the core of these systems due to their favorable superparamagnetic properties, which allow them to be dispersed in a solution for efficient binding and then collected using an external magnetic field without retaining residual magnetism. This application note details the use of advanced substrates, with a focus on silica-coated iron oxide nanoparticles (Fe₃O₄@SiO₂), for automated nucleic acid extraction from stool samples, providing researchers with detailed protocols and performance data to enhance their diagnostic and research workflows.
Material Properties and Characterization
Core Magnetic Particle Synthesis and Functionalization
The performance of magnetic nanoparticles in nucleic acid extraction is fundamentally governed by their synthesis and surface functionalization. The core Fe₃O₄ nanoparticles are typically synthesized via methods such as the polyol process or co-precipitation, which allow for control over morphology, size, and magnetic properties [27] [28]. A critical advancement for application in stool samples is the surface coating of these magnetic cores, which enhances stability, prevents agglomeration, and provides functional groups for nucleic acid binding.
Key coatings include:
- Silica (SiO₂): Coating via the Stöber method introduces a surface rich in silanol groups that facilitate DNA binding in the presence of chaotropic salts [27] [29]. The silica coating creates a protective layer that shields the magnetic core from the harsh chemical environment sometimes present in stool lysates.
- Polyethyleneimine (PEI): A polymer coating that provides a high density of positively charged amine groups, enabling strong electrostatic interaction with the negatively charged phosphate backbone of nucleic acids. A comparative study found Fe₃O₄@PEI to be the most efficient nano-sorbent for dsDNA extraction [30].
- Oleic Acid (OA): Often used as an initial surfactant coating to stabilize naked Fe₃O4 nanoparticles and provide a foundation for subsequent silica coating (Fe₃O₄@OA@SiO₂) [29].
Table 1: Characteristics of Functionalized Iron Oxide Nanoparticles for Nucleic Acid Extraction
| Material/Coating | Core Synthesis Method | Key Coating Properties | Primary Interaction with Nucleic Acids | Key Advantage for Stool Samples |
|---|---|---|---|---|
| Fe₃O₄@SiO₂ | Polyol, Co-precipitation | Silanol groups, hydrophilic | Electrostatic, cation-bridging with chaotropes | Robustness against inhibitors, high purity yields [27] [29] |
| Fe₃O₄@PEI | Co-precipitation | High-density amine groups, cationic | Direct electrostatic binding | Highest DNA adsorption efficiency, works in low-EDTA buffers [30] |
| Fe₃O₄@OA@SiO₂ | Ultrasound-assisted co-precipitation | Bilayer: hydrophobic OA + hydrophilic SiO₂ | Silanol-mediated (as above) | Enhanced stability and nucleic acid absorption vs. OA-only [29] |
| Gold-coated | Not Specified | Gold surface chemistry | Not Specified | Evaluated for DNA extraction, less efficient than PEI [30] |
Magnetic and Physical Properties
The synthesized nanoparticles must be characterized to ensure quality and performance. Key analyses include:
- Transmission Electron Microscopy (TEM): Used to confirm particle size and morphology. For instance, Fe₃O₄@OA@SiO₂ particles showed a mean diameter of 106 nm with a clear core-shell structure [29].
- Vibrating Sample Magnetometry (VSM): Confirms superparamagnetic behavior, characterized by high magnetic saturation and no magnetic remanence. This ensures particles will not aggregate after the magnetic field is removed [29].
- Fourier-Transform Infrared (FTIR) Spectroscopy: Verifies the success of surface functionalization by identifying characteristic chemical bonds, such as the Si-O-Si stretch for silica coatings or the carbonyl stretch for oleic acid [27] [29].
Performance Evaluation in Nucleic Acid Extraction
Quantitative Extraction Efficiency
The efficacy of different functionalized MNPs has been quantitatively evaluated in multiple studies. Silica-coated MNPs have demonstrated high efficiency, with one study showing that 0.5 mg of silica-coated Fe₃O₄ particles could yield an average of 2.88 μg of nucleic acids [27]. Furthermore, the purity and integrity of the isolated nucleic acids are suitable for demanding downstream applications, including PCR and next-generation sequencing.
In a comprehensive comparative evaluation of Fe₃O₄-based sorbents with different coatings (PEI, gold, silica, and graphene oxide), Fe₃O₄@PEI MNPs were identified as the most efficient nano-sorbents for dsDNA extraction [30]. The study also highlighted that optimizing the extraction buffer, specifically using a medium containing 0.1 mM EDTA, improved the validity of spectroscopic DNA recovery determination by minimizing Fe³⁺ stripping.
When isolating DNA from complex samples like cyanobacteria (Arthrospira platensis) and animal blood, Fe₃O₄@OA@SiO₂ produced 1.2 and 1.6 times greater DNA yield, respectively, compared to Fe₃O₄@OA, underscoring the advantage of the silica coating for nucleic acid adsorption [29].
Comparative Performance in Automated Systems and Challenging Samples
For automated, high-throughput diagnostics, consistency across production batches is paramount. Scaling up the synthesis of TEOS-modified magnetic particles from 1 L to 5 L has been shown to be feasible, with minimal batch-to-batch variability in DNA extraction performance, ensuring reproducible results in clinical settings [28].
For challenging stool samples, inhibitor removal is critical. A study on rotavirus RNA detection in stool compared six extraction methods and found that extracts from the MagNA Pure Compact system (which utilizes magnetic particle technology) provided the most consistent results in qRT-PCR and conventional RT-PCR, with effective removal of inhibitors [7]. Another broader study on respiratory pathogens found that while all evaluated automated magnetic-bead systems recovered nucleic acids effectively, their performance varied in a pathogen-specific manner, suggesting that the choice of system can be optimized for particular targets [31].
Table 2: Performance Summary of Magnetic Particle-Based Extraction Methods
| Extraction System / Material | Sample Type | Reported Performance Metric | Key Finding |
|---|---|---|---|
| Fe₃O₄@PEI MNPs | Model DNA systems | DNA Adsorption Efficiency | Most efficient nano-sorbent among coatings tested [30] |
| Silica-coated Fe₃O₄ | Model DNA/RNA systems | Nucleic Acid Yield | 0.5 mg particles yielded 2.88 (2.67–3.08) μg of nucleic acids [27] |
| Fe₃O₄@OA@SiO₂ | Arthrospira platensis, Blood | DNA Yield (Relative Increase) | 1.6x greater DNA from blood vs. Fe₃O₄@OA [29] |
| MagNA Pure Compact | Stool (Rotavirus) | PCR Consistency & Inhibitor Removal | Most consistent qRT-PCR results; effective inhibitor removal [7] |
| Scaled-up TEOS-MPs | Plasma (Viral NA) | Batch-to-Batch Reproducibility | Minimal variability in DNA extraction with scaled-up synthesis [28] |
Detailed Experimental Protocols
Protocol 1: Automated Nucleic Acid Extraction from Stool Samples Using Functionalized Magnetic Particles
This protocol is adapted for use with an automated nucleic acid extractor (e.g., MagNA Pure, KingFisher) and functionalized silica-coated magnetic particles.
I. Reagents and Materials
- Lysis Buffer: 4-6 M guanidine hydrochloride, 10 mM EDTA, 20 mM Tris-HCl (pH 6.6), 1% Triton X-100.
- Wash Buffer 1: 4 M guanidine hydrochloride, 20 mM EDTA, 10 mM Tris-HCl (pH 6.6), and 50% ethanol.
- Wash Buffer 2: 70% ethanol, 10 mM KCl, 2 mM EDTA, 10 mM Tris-HCl (pH 7.5).
- Elution Buffer: Nuclease-free water or 10 mM Tris-HCl (pH 8.5).
- Proteinase K (optional, for enhanced lysis).
- Functionalized Silica Magnetic Particles (e.g., Fe₃O₄@SiO₂ suspension).
II. Equipment
- Automated Magnetic Particle Processor (e.g., MagNA Pure Compact, KingFisher Flex).
- Microcentrifuge tubes or deep-well plates compatible with the automated system.
- Vortex mixer and heating block (for pre-processing).
- Magnetic stand (if manual steps are involved).
III. Procedure
- Sample Preparation and Lysis:
- Using a device like the Easy Stool Extraction Device, homogenize 10-20 mg of stool sample in 1 mL of universal extraction buffer [32]. Alternatively, suspend the stool sample directly in Lysis Buffer.
- Incubate the mixture at 65-70°C for 10-15 minutes. If necessary, add Proteinase K and incubate at 56°C for an additional 10-20 minutes to digest proteins.
- Centrifuge the lysate at >12,000 × g for 2 minutes to pellet insoluble debris.
Nucleic Acid Binding:
- Transfer the clarified supernatant to a new tube or the well of a processing plate.
- Add a predetermined volume of the functionalized magnetic particle suspension (e.g., equivalent to 0.5-1 mg of particles) to the lysate.
- Mix thoroughly by pipetting or vortexing and incubate at room temperature for 5-10 minutes. During this period, nucleic acids bind to the particles' surface.
Magnetic Separation and Washing:
- Engage the magnetic field in the automated system to capture the particle-nucleic acid complex. Discard the supernatant.
- With the magnetic field engaged, wash the particles with 500 μL of Wash Buffer 1. Fully resuspend the pellet to ensure complete removal of contaminants. Discard the flow-through.
- Repeat the wash step with 500 μL of Wash Buffer 2.
- Perform a final quick wash with 100% ethanol or allow a brief air-dry cycle (~2-5 minutes) to evaporate residual ethanol.
Elution:
- Resuspend the washed magnetic particles in 50-100 μL of Elution Buffer.
- Incubate at 65-70°C for 5 minutes to facilitate the release of nucleic acids from the particles.
- Engage the magnetic field once more and transfer the eluate containing the purified nucleic acids to a clean tube.
- The extracted nucleic acids are now ready for downstream applications. Store at -20°C or -80°C for long-term preservation.
Protocol 2: Synthesis of Silica-Coated Iron Oxide Nanoparticles (Fe₃O₄@OA@SiO₂)
This two-step protocol describes the synthesis of core-shell nanoparticles suitable for nucleic acid extraction [29].
I. Reagents
- Ferric chloride hexahydrate (FeCl₃·6H₂O)
- Ferrous chloride tetrahydrate (FeCl₂·4H₂O)
- Sodium hydroxide (NaOH)
- Oleic Acid (OA)
- Absolute Ethanol
- Tetraethyl orthosilicate (TEOS)
- Ammonia solution (NH₃)
- Double-distilled water (ddH₂O)
II. Equipment
- Ultrasonic bath
- Round-bottom flask
- Centrifuge
- Magnetic stirrer
- Drying oven
III. Procedure
- Synthesis of Oleic Acid-Coated Nanoparticles (Fe₃O₄@OA):
- Dissolve 1.03 g of FeCl₃·6H₂O and 0.01 mol of FeCl₂·4H₂O in 40 mL of ddH₂O in a round-bottom flask.
- Sonicate the mixture at 50°C for 20 minutes until the salts are fully dissolved.
- Slowly add 40 mL of 0.8 M NaOH solution dropwise (approximately 1 droplet every 2 seconds) under continuous sonication.
- Continue sonication for 1 hour after complete NaOH addition to form a black precipitate of Fe₃O₄.
- Add 20 mL of oleic acid to the mixture and sonicate for an additional 1 hour to coat the particles.
- Recover the Fe₃O₄@OA nanoparticles by centrifugation at 10,000 rpm for 30 minutes at 4°C. Wash the pellet with absolute ethanol five times to remove unreacted precursors.
- Dry the final product at 50°C for 15 hours.
- Silica Coating via the Stöber Method (Fe₃O₄@OA@SiO₂):
- Disperse 40 mg of the synthesized Fe₃O₄@OA in 16.8 mL of ddH₂O by sonication for 20 minutes.
- To this dispersion, add 64 mL of ethanol, 4 mL of ammonia, and 4 mL of TEOS under continuous ultrasonic vibration.
- Allow the reaction to proceed for 4-6 hours to form the silica shell.
- Recover the Fe₃O₄@OA@SiO₂ nanoparticles by centrifugation at 10,000 rpm for 30 minutes at 4°C. Wash the pellet with absolute ethanol five times.
- Dry the final silica-coated particles at 50°C for 15 hours. The nanoparticles can be stored as a dry powder or suspended in a stable buffer for future use.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents and Materials for Magnetic Particle-Based NA Extraction
| Reagent/Material | Function/Description | Application Note |
|---|---|---|
| Chaotropic Salts (e.g., Guanidine HCl) | Denature proteins & facilitate NA binding to silica surface by dehydrating the NA backbone. | Critical for efficient binding in silica-based protocols; concentration typically 4-6 M [31]. |
| Functionalized Magnetic Particles (e.g., Fe₃O₄@SiO₂) | Solid-phase substrate for NA binding, washing, and elution. | Core innovation; particle size, coating, and magnetization are key performance factors [27] [28]. |
| Proteinase K | Broad-spectrum serine protease for digesting proteins and nucleases. | Essential for stool samples to degrade proteinaceous inhibitors and release NA from complex matrices [33]. |
| Ethanol-based Wash Buffers | Remove salts, solvents, and other contaminants from the particle-NA complex. | Ensures high purity of final eluate; residual ethanol must be evaporated to prevent inhibition of downstream assays [7]. |
| Low-Ionic-Strength Elution Buffer (e.g., Tris, Water) | Disrupts NA-particle interaction by rehydrating the NA backbone. | Elution at elevated temperature (65-70°C) often increases final yield [33]. |
| Easy Stool Extraction Device | Standardizes collection and initial homogenization of stool samples. | Ensures consistent starting material (e.g., 15 mg stool in 1.5 mL buffer), improving reproducibility [32]. |
The pursuit of efficiency, reproducibility, and minimization of contamination in molecular biology has driven the development of integrated nucleic acid extraction workflows. Traditional methods involving multiple tube transfers and manual handling pose significant risks of sample loss, cross-contamination, and procedural variability. This is particularly relevant for complex sample matrices like stool, which contain numerous PCR inhibitors that can compromise downstream applications [12]. Single-tube protocols address these challenges by consolidating lysis, purification, and often elution into a single reaction vessel, offering substantial improvements in workflow efficiency and data reliability for researchers and drug development professionals working with automated nucleic acid extraction from stool samples.
This application note details several established and emerging single-tube methodologies, providing quantitative performance comparisons and detailed experimental protocols to facilitate their implementation in automated stool sample processing.
Single-Tube Methodologies and Performance Data
Different technological approaches have been successfully employed to create integrated nucleic acid extraction workflows. The table below summarizes the core characteristics and performance metrics of several key methods.
Table 1: Comparison of Single-Tube Nucleic Acid Extraction Methods
| Method Name | Core Technology | Sample Types Demonstrated | Processing Time | Key Performance Metrics | Reference |
|---|---|---|---|---|---|
| PurAmp | Chaotropic lysis with sequential dilution | Single mouse embryos, blastomeres | Rapid (specific time not given) | Sensitive to single molecules; quantitative DNA/RNA recovery | [34] |
| SHIFT-SP | Magnetic silica beads with tip-based mixing | Mycobacterium smegmatis DNA, whole blood | 6–7 minutes | ~96% binding efficiency; nearly complete NA elution | [16] |
| AOM Tubes | Aluminum oxide membrane filtration | Herpes Simplex Virus in CSF | ~15-20 minutes (incl. vacuum steps) | 100% concordance with reference method | [35] |
| HTP Centrifugal Microfluidic | Centrifugal microfluidics with silica-based purification | Clinical samples (10 simultaneously) | < 20 minutes for 10 samples | High-quality RNA for downstream RT-PCR | [36] |
These methods showcase a trend toward miniaturization, automation, and significantly reduced processing times, all while maintaining or improving the yield and purity of extracted nucleic acids compared to multi-step protocols.
Detailed Experimental Protocols
The PurAmp Single-Tube Protocol for Sensitive Applications
The PurAmp method is designed for maximum recovery in minute samples, eliminating purification steps through volumetric dilution of chaotropes [34].
Materials:
- Lysis Buffer: 5 M Guanidine Isothiocyanate (GITC)
- Enzymes: Proteinase K (optional for DNA-only extraction)
- Equipment: Thermal cycler with real-time PCR detection
Procedure:
- Lysis: Transfer a single cell or small tissue sample (e.g., a mouse embryo) into a thin-walled PCR tube containing 5-10 µL of GITC lysis buffer.
- Incubation: Incubate the tube at room temperature for 5 minutes to ensure complete lysis and protein denaturation. The sample can be stored dry at this stage if needed.
- Neutralization and RT-PCR: Directly add the complete RT-PCR master mix to the same tube. The master mix volume must be sufficient to dilute the GITC concentration to a level that does not inhibit enzymatic activity (typically a >20-fold dilution).
- Amplification: Perform reverse transcription followed by real-time PCR in the same tube without any transfer steps.
Critical Considerations: This protocol is highly dependent on the initial sample-to-lysis buffer volume ratio and the subsequent dilution factor. The high dilution factor required can be a limitation for some downstream applications.
The SHIFT-SP High-Yield Magnetic Bead Protocol
This protocol optimizes binding and elution for rapid, high-efficiency extraction using magnetic silica beads [16].
Materials:
- Lysis/Binding Buffer (LBB): Guanidine hydrochloride, Triton X-100, pH adjusted to 4.1
- Magnetic Silica Beads
- Wash Buffers: 100 mmol/L NaCl solution
- Elution Buffer: 1X TE Buffer or nuclease-free water
- Equipment: Magnetic stand, pipettes
Procedure:
- Lysis and Binding:
- Combine the sample (e.g., 100 µL of bacterial culture lysate) with an equal volume of LBB (pH 4.1) in a tube.
- Add 30-50 µL of magnetic silica bead suspension.
- Perform "tip-based" binding: Repeatedly aspirate and dispense the mixture with a pipette for 1-2 minutes at 62°C. This ensures rapid and efficient exposure of nucleic acids to the beads.
- Bead Capture: Place the tube on a magnetic stand until the solution clears. Carefully remove and discard the supernatant.
- Washing:
- With the tube on the magnetic stand, add 200 µL of 100 mmol/L NaCl wash buffer. Disperse the beads by flicking the tube or brief vortexing.
- Capture the beads and discard the supernatant.
- Repeat the wash once with 200 µL of nuclease-free water.
- Elution: Remove the tube from the magnetic stand. Add 20-50 µL of Elution Buffer and resuspend the beads. Incubate at 70°C for 1 minute to facilitate high-yield elution. Capture the beads and transfer the eluate containing purified nucleic acids to a new tube.
Automated Single-Tube Workflow for Stool Samples
For complex matrices like stool, commercial automated systems offer robust, integrated solutions [12].
Materials:
- Kit: EZ2 PowerFecal Pro DNA/RNA Kit (QIAGEN)
- Equipment: EZ2 Connect instrument
- Reagents: Phenol:chloroform:isoamyl alcohol, Inhibitor Removal Technology (IRT) buffer
Procedure:
- Lysis and Homogenization: Weigh 50-100 mg of stool and place it in a tube prefilled with lysis buffer and beads. Securely close the tube and homogenize using a vortex adapter or a benchtop homogenizer like the Tissuelyser III.
- Inhibitor Removal: Centrifuge the lysate briefly. Transfer the supernatant to a new tube and add phenol:chloroform:isoamyl alcohol. Vortex and centrifuge to separate phases. The proprietary IRT chemistry in the buffer further removes PCR inhibitors.
- Automated Processing: Transfer the aqueous upper phase to a pre-filled cartridge and load it onto the EZ2 Connect instrument.
- Hands-Off Extraction: Select the desired protocol (DNA-only, RNA-only, or Total Nucleic Acids). The instrument automatically performs all subsequent steps, including optional DNase/RNase digestion, binding, washing, and elution, in a closed system.
- Recovery: Retrieve the eluate containing high-purity nucleic acids from the instrument, ready for downstream applications.
Workflow Integration and Visualization
The following diagram illustrates the logical sequence and decision points in a generalized, automated single-tube workflow suitable for nucleic acid extraction from stool samples.
The Scientist's Toolkit: Essential Research Reagent Solutions
Successful implementation of single-tube protocols relies on specific reagents and tools designed for integrated workflows.
Table 2: Key Research Reagent Solutions for Single-Tube Workflows
| Reagent/Tool | Function in Workflow | Example Use-Case |
|---|---|---|
| Chaotropic Salts (e.g., GITC, Guanidine HCl) | Denature proteins and nucleases; facilitate binding of nucleic acids to silica surfaces. | Cell lysis and nuclease inactivation in the initial phase (PurAmp, SHIFT-SP) [34] [16]. |
| Silica-Magnetic Beads | Solid-phase matrix for nucleic acid binding; enables separation via a magnetic field without centrifugation. | Automated nucleic acid capture and washing in platforms like the KingFisher (MagMAX kits) and EZ2 Connect [12] [21]. |
| Inhibitor Removal Technology (IRT) Buffers | Proprietary chemistries designed to adsorb and remove specific PCR inhibitors from complex samples. | Critical for obtaining amplifiable DNA/RNA from inhibitor-rich stool samples [12]. |
| Alternative Matrices (e.g., Aluminum Oxide Membrane - AOM) | Porous filter that captures nucleic acids under vacuum or pressure, integrated into a PCR-tube format. | Single-tube extraction, amplification, and detection of viral pathogens from CSF [35]. |
| Pre-filled Reagent Cartridges | Ensure reagent consistency, reduce pipetting errors, and streamline the setup of automated systems. | Used in automated platforms like the EZ2 Connect for processing stool samples with the PowerFecal Pro kit [12]. |
Integrated single-tube protocols represent a significant advancement in nucleic acid extraction technology. By consolidating lysis, purification, and elution into a single vessel or a fully automated workflow, these methods enhance throughput, improve reproducibility, and minimize the risk of contamination. For research and drug development programs focused on the microbiome and other stool-based analyses, adopting these streamlined workflows is crucial for generating robust, reliable, and high-quality molecular data.
The reliability of downstream molecular analyses, from quantitative PCR (qPCR) to next-generation sequencing (NGS), is fundamentally dependent on the quality and integrity of the isolated nucleic acids. This is particularly challenging with stool samples, which contain complex mixtures of undigested food, gut microbiota, host cells, and potent PCR inhibitors like bile salts and complex polysaccharides [37] [38]. Automated nucleic acid extraction systems have therefore become indispensable in modern laboratories, enabling high-throughput, reproducible purification of DNA and RNA while minimizing manual labor and cross-contamination risk [39] [40].
This application note provides a performance analysis of leading commercial automated nucleic acid extraction platforms, with a specific focus on their application for stool samples. We summarize key performance metrics and provide a detailed protocol optimized for challenging stool matrices.
The global market for automated nucleic acid extraction is characterized by rapid technological innovation and growth, driven by demands in molecular diagnostics, genomics research, and personalized medicine [41] [39]. North America currently holds the dominant market share, propelled by well-established healthcare infrastructure, significant investments in genomic research, and early adoption of automated technologies [39] [42]. The market is segmented by product type (instruments, kits, consumables), technology (magnetic bead-based, column-based), and end-user (hospitals, diagnostic centers, pharmaceutical and biotechnology companies, academic research institutes) [40] [43].
Magnetic bead-based technology has emerged as the dominant and fastest-growing segment due to its scalability, high yield, efficiency with diverse sample types, and low contamination risk, making it particularly suitable for automated, high-throughput workflows [39] [42].
Table 1: Key Commercial Automated Nucleic Acid Extraction Platforms
| Platform Name | Key Technology | Throughput (Samples per Run) | Primary Application Focus | Notable Features |
|---|---|---|---|---|
| VERSA NAP (Aurora Biomed) | Magnetic Beads | 1-96 [40] | DNA/RNA Extraction & Purification | Can process 1-96 samples simultaneously; uses plate grippers and transporters [40]. |
| SAW-48 (Jiangsu Bioperfectus) | Magnetic Beads | Up to 48 [43] | In-Vitro Diagnostics | Integrates sample loading, nucleic acid purification, and PCR setup in one instrument [43]. |
| Monarch Mag Viral DNA/RNA Extraction Kit (New England Biolabs) | Magnetic Beads | High-throughput compatible [42] | Viral Nucleic Acid Extraction | Optimized for sensitive detection of minute viral quantities from saliva, respiratory swabs, wastewater [42]. |
| Systems using SmartLid (Alpha Laboratories/ProtonDx) | Magnetic Beads | N/A | Viral DNA/RNA Extraction | Simplified process with a magnetic key and lid system for lysis, wash, and elution [42]. |
| sbeadex Lightning (LGC Biosearch Technologies) | Magnetic Beads | High-throughput [42] | Plant & Animal DNA Extraction | Rapid 5-minute extraction workflow; increases sample throughput up to 10x [42]. |
Experimental Protocol: Automated Nucleic Acid Extraction from Stool Samples
Pre-Analytical Sample Preparation
Proper sample collection and preparation are critical for success.
- Sample Collection: Use a sterile, leak-proof stool collection container, often with an attached spoon [44]. For comprehensive analysis, collect samples on two separate days (at least 12 hours apart) [37].
- Patient Preparation: Patients should discontinue probiotics for three days prior to collection, as they can skew microbial population results. Digestive enzymes, antacids, iron supplements, high-dose vitaminC (>250 mg), aspirin, anti-inflammatories, and large amounts of meat should be avoided 48 hours before collection [37].
- Storage and Transport: Store samples at recommended temperatures and ensure they reach the processing lab within the required timeframe (e.g., within seven days of first collection) [37]. Post samples early in the week (Monday-Wednesday) to avoid weekend transit delays [38].
- Homogenization: Create a homogeneous stool suspension by weighing an appropriate amount of stool (e.g., 100-200 mg) and diluting it in a specific buffer provided in the extraction kit. Vortex thoroughly to ensure a consistent mixture.
Automated Extraction Workflow using Magnetic Bead Technology
The following protocol outlines the general steps for a magnetic bead-based automated extraction, which is the prevailing technology [39] [42].
Diagram: Automated magnetic bead-based nucleic acid extraction workflow.
- Step 1: Lysis and Binding. The homogenized stool sample is transferred to a deep-well plate. The automated system adds a lysis buffer containing chaotropic salts (e.g., guanidinium thiocyanate) and proteinase K to break down cell walls, viral envelopes, and nucleases, releasing nucleic acids into solution. Magnetic beads, which bind nucleic acids in the presence of a high-salt concentration and ethanol, are added. The mixture is incubated with vigorous shaking to facilitate binding [42].
- Step 2: Magnetic Separation. The instrument engages a magnetic field to immobilize the bead-nucleic acid complexes against the wall of the plate. The supernatant, containing stool debris, proteins, and inhibitors, is automatically removed and discarded.
- Step 3: Washing. While the magnetic field is still applied, the system sequentially adds and removes two or more wash buffers (typically an ethanol-based wash buffer followed by a final alcohol wash). These steps remove salts, metabolites, and other impurities without eluting the bound nucleic acids. Some advanced systems, like those using the SmartLid technology or the sbeadex Lightning kit, streamline this into a more efficient process [42].
- Step 4: Elution. The magnetic field is disengaged. A low-salt elution buffer (e.g., Tris-EDTA buffer or nuclease-free water) is added to the dried magnetic beads. The mixture is resuspended and incubated, causing the nucleic acids to dissociate from the beads. The magnetic field is reapplied, and the eluate containing the purified nucleic acids is automatically transferred to a fresh output plate, ready for downstream applications.
Post-Extraction Quality Control and Downstream Analysis
- Quality Control: Assess the concentration and purity of eluted DNA/RNA using spectrophotometry (e.g., Nanodrop) or fluorometry (e.g., Qubit). Analyze integrity via agarose gel electrophoresis or automated systems (e.g., Bioanalyzer).
- Downstream Application: The purified nucleic acids can be used for various applications, including PCR/qPCR for pathogen detection, 16S rRNA sequencing for microbiome analysis, shotgun metagenomics, or transcriptomic studies of the gut microbiome.
The Scientist's Toolkit: Essential Reagents and Materials
A successful automated extraction relies on a suite of optimized reagents and consumables.
Table 2: Essential Research Reagent Solutions for Automated Extraction
| Reagent/Material | Function | Key Considerations |
|---|---|---|
| Lysis Buffer | Disrupts cells and viral particles; inactivates nucleases; provides ideal chemical conditions for nucleic acid binding to beads. | Often contains chaotropic salts and detergents. Must be optimized for robust lysis of diverse microbes in stool (Gram+, Gram-, spores). |
| Binding Beads | Silica-coated magnetic particles that selectively bind nucleic acids in the presence of chaotropic salts and alcohol. | The core of the technology. Size, coating, and uniformity impact yield, purity, and consistency [39]. |
| Wash Buffers | Remove proteins, salts, and other contaminants from the bead-nucleic acid complex without causing elution. | Typically contain ethanol or isopropanol. Stringent vs. gentle wash buffers may be used sequentially. |
| Elution Buffer | A low-ionic-strength solution (e.g., TE buffer or water) that destabilizes the nucleic acid-bead interaction, releasing pure nucleic acids into solution. | Volume and pH affect final concentration and stability of the eluted nucleic acids. |
| Proteinase K | A broad-spectrum serine protease that digests denatured proteins and nucleases, critical for efficient lysis and protecting nucleic acids. | Essential for breaking down tough structures in stool and inactivating RNases/DNases. |
| Internal Control | A non-native nucleic acid sequence added to the sample at the beginning of lysis. | Monitors extraction efficiency and identifies the presence of PCR inhibitors in the final eluate. |
| Deep-Well Plates & Tips | Disposable plasticware designed for use with the automated liquid handling system. | Must be compatible with the specific platform and free of DNase/RNase. |
Automated nucleic acid extraction systems are pivotal for enabling robust, high-throughput molecular analysis of complex stool samples. The trend is firmly centered on magnetic bead-based technology, which offers superior efficiency, scalability, and minimal cross-contamination [39] [42]. When selecting a platform, researchers must balance throughput, cost, and the specific demands of their stool-based research questions. The provided protocol and toolkit offer a foundation for implementing these powerful technologies, ensuring the generation of high-quality nucleic acids that are the bedrock of reliable and actionable genomic data.
The Simple One-Step (SOS) Stool Processing Method represents a significant advancement for molecular testing in clinical and research settings, particularly for the diagnosis of pulmonary tuberculosis (TB) using the Xpert MTB/RIF Ultra (Xpert-Ultra) assay [45]. Its development addresses a critical diagnostic challenge: obtaining bacteriological confirmation of TB in children, who often cannot produce sputum [46]. The World Health Organization (WHO) and the Global Laboratory Initiative (GLI) now recommend stool as a primary, non-invasive diagnostic specimen for TB in children and have endorsed the SOS method as one of two preferred processing techniques [45] [46].
The method's primary strength in a research context focused on automated nucleic acid extraction is its simplicity and minimal requirement for additional equipment. The SOS method utilizes a single release-sedimentation step where heavy debris settles by gravity, allowing target bacteria to remain in the supernatant [46]. This aligns stool processing workflows closely with established sputum testing protocols for the GeneXpert system, facilitating implementation in routine clinical and research laboratories without the need for centrifuges or complex instrumentation [45] [47]. Understanding and standardizing this method is a crucial step for integrating robust, high-throughput stool processing into broader research on automated nucleic acid extraction from complex biological matrices.
The Optimized SOS Stool Protocol
The following section details the standardized operating procedure for the SOS method, incorporating key optimizations from robustness testing.
Materials and Reagents
Table 1: Essential Research Reagent Solutions for SOS Stool Processing
| Item | Function/Description | Specification/Note |
|---|---|---|
| Xpert MTB/RIF Ultra Assay | Molecular cartridge for detection of M. tuberculosis and rifampin resistance. | Includes Sample Reagent (XSR) bottle [48]. |
| Xpert Sample Reagent (XSR) | Chemical reagent for sample treatment; lyses organisms and inactivates contaminants [48]. | Supplied with Xpert Ultra assay [48]. |
| Stool Collection Container | Wide-mouth, leak-proof container for sample collection. | Should allow for collection of at least 30g of stool [45]. |
| Applicator Stick | For measuring and transferring stool sample. | Sterile, single-use [48]. |
| Timer | For standardizing incubation and sedimentation steps. | - |
| Transfer Pipette | For transferring supernatant to Xpert cartridge. | - |
Step-by-Step Workflow
The workflow for the SOS Stool Method, from collection to analysis, is designed to be simple and robust.
- Sample Collection: Collect a fresh stool specimen in a clean, dry, wide-mouth container. A minimum of 30 grams is recommended to allow for multiple tests or repeats [45].
- Weigh Stool Sample: Using an applicator stick, weigh and transfer 0.3 to 0.8 grams of stool into the 8 ml XSR bottle. Adhering to this optimized range is critical, as using more than 0.8g significantly increases the rate of processing errors [46].
- Initial Mixing: Close the XSR bottle cap securely and shake it vigorously for 30 seconds to homogenize the stool with the reagent [48].
- Incubation: Let the mixture stand at room temperature for 10 minutes [48].
- Second Mixing: Shake the bottle vigorously again for 30 seconds [48].
- Sedimentation: Allow the mixture to sediment undisturbed at room temperature for 10 minutes. During this step, heavy particles settle, leaving the target organisms in the supernatant [46] [48].
- Supernatant Transfer: Using a transfer pipette, carefully aspirate 2 ml of the supernatant without disturbing the sediment pellet [48].
- Cartridge Loading: Transfer the 2 ml of supernatant directly into the Xpert MTB/RIF Ultra cartridge [48].
- Instrument Operation: Load the cartridge into the GeneXpert instrument and start the test run as per the manufacturer's instructions.
Experimental Validation and Robustness Data
Robustness testing of the SOS method has provided critical quantitative data to define optimal and acceptable parameters for its use in research and clinical workflows.
Impact of Stool Sample Volume
Experiments evaluating different stool sample masses revealed a direct correlation between increased mass and error rates, leading to a key protocol optimization.
Table 2: Effect of Stool Sample Volume on Xpert-Ultra Test Results
| Stool Mass (g) | MTB Positivity Rate | Rate of Processing Errors | Recommendation |
|---|---|---|---|
| 0.3 | Unchanged | 3.7% | Optimal lower limit |
| 0.8 | Unchanged | ~5% | Original recommended mass |
| >0.8 | Unchanged | >20% (Significant increase) | Avoid; sharply increases errors |
The data demonstrates that while the MTB detection sensitivity remains stable across a range of masses, the reliability of the test is compromised when the stool mass exceeds 0.8 grams. The rate of processing errors rose significantly to 20.2% at 1.2 grams of stool [46]. Consequently, the protocol was adjusted to recommend a range of 0.3 to 0.8 grams, providing flexibility while ensuring result reliability [46].
Stool Sample Storage Conditions
Understanding pre-processing storage conditions is vital for practical implementation, especially when immediate testing is not feasible.
Table 3: Impact of Storage Time and Temperature on Stool Specimens
| Storage Temperature | Maximum Storage Duration | Effect on Xpert-Ultra Results |
|---|---|---|
| Room Temp (20-22°C) | Up to 3 days | No significant loss of MTB sensitivity [45]. |
| Refrigerated (2-8°C) | Up to 5 days | No significant loss of MTB sensitivity [45]. |
| Elevated Temp (37°C) | - | Not recommended; potential for sensitivity loss [45]. |
| Stool-XSR Mixture | ≤ 12 hours at Room Temp | Maintains reliability [46]. |
| Stool-XSR Mixture | > 12 hours at Room Temp | Significantly increases error rates [46]. |
The experiments indicate that stool specimens can be stored for several days without refrigeration without degrading MTB detection sensitivity. However, once the stool is mixed with the XSR reagent, the test should ideally be completed within 12 hours to prevent a rise in indeterminate results [46].
Experimental Protocol for Robustness Testing
The following methodology was used to generate the critical data on stool volume and storage robustness [45] [46]:
- Sample Preparation: Stool specimens from participants with bacteriologically confirmed TB were used. Multiple aliquots were taken from each homogenized, known positive stool specimen.
- Variable Testing:
- For volume testing, aliquots of different masses (e.g., 0.3g, 0.8g, 1.2g) were processed according to the standard SOS protocol and run on Xpert-Ultra.
- For storage condition testing, aliquots of a positive stool were stored at different temperatures (refrigerated, room temperature, 37°C) and for different durations (2, 3, 5, and 10 days). After storage, each aliquot was processed and tested.
- Data Analysis: The primary outcomes measured were the MTB positivity rate (sensitivity) and the rate of Xpert-Ultra processing errors (invalid, error, no result). Results for each test condition were compared to the baseline result from a freshly processed 0.8g sample.
The structure of these robustness experiments demonstrates a systematic approach to validating a clinical workflow.
Integration with Automated Nucleic Acid Extraction Research
While the SOS method itself is a manual sample preparation technique, its principles and requirements inform and intersect with the field of automated nucleic acid extraction in key ways.
- Workflow Synergy: The SOS method serves as a critical "front-end" sample preparation step, transforming a complex raw stool sample into a clarified supernatant compatible with downstream automated systems. Automated extractors, such as the KingFisher Apex or Maxwell RSC, are highly effective at purifying nucleic acids from liquid samples but can be hampered by the inhibitors and particulate matter in raw stool [5]. The SOS method effectively bridges this gap.
- Inhibitor Removal: A major challenge in stool-based molecular testing is the presence of PCR inhibitors. The SOS method's use of XSR and the sedimentation step is designed to mitigate this [46]. This goal is paralleled in commercial automated extraction kits for stool, such as the MagMAX Microbiome Ultra Kit and QIAGEN EZ2 PowerFecal Pro Kit, which incorporate specialized inhibitor removal technologies and mandatory bead-beating steps for efficient microbial lysis [5] [12].
- Comparative Performance: Studies comparing automated nucleic acid extraction systems for stool highlight factors critical for research reproducibility. For instance, the inclusion of bead-beating (mechanical lysis) was found to be essential for the effective lysis of Gram-positive bacteria and for providing a greater representation of the microbial community in stool, impacting downstream sequencing results [5]. This is a consideration for researchers who may use the SOS method for initial screening and then perform automated extraction for broader microbiome or metagenomic analysis.
Table 4: Comparison of Automated Nucleic Acid Extraction Systems for Stool
| Extraction System | Technology | Bead-Beating Required | Key Considerations for Stool |
|---|---|---|---|
| KingFisher Apex | Magnetic Bead-Based | Yes [5] | High throughput (1-96 samples); effective for diverse bacterial lysis [5]. |
| Maxwell RSC | Magnetic Bead-Based | Yes/No (Separate step) [5] | Mid-throughput (1-16 samples); bead-beating improves Gram-positive yield [5]. |
| GenePure Pro | Magnetic Bead-Based | Yes/No (Separate step) [5] | Semi-automatic; lower yield without bead-beating [5]. |
| EZ2 Connect with PowerFecal Pro | Magnetic Bead-Based | Yes (per protocol) [12] | Integrated inhibitor removal technology; pre-filled cartridges minimize error [12]. |
The Simple One-Step stool processing method is a validated, robust, and simple-to-implement protocol that standardizes the use of stool for molecular detection of M. tuberculosis. Its optimization, characterized by the defined stool mass range of 0.3 to 0.8 grams and flexible storage conditions of up to 3 days at room temperature, provides clear guidelines for reliable integration into clinical and research workflows. For the field of automated nucleic acid extraction, the SOS method stands as a complementary and enabling sample preparation technique, handling the initial critical steps of sample homogenization and inhibitor removal to ensure that downstream automated systems receive a compatible and high-quality input. Its adoption facilitates a more accessible and scalable path to bacteriological confirmation of TB, particularly in resource-limited settings.
Solving Common Problems and Maximizing Extraction Efficiency and Yield
The success of polymerase chain reaction (PCR) in modern molecular biology, particularly in applications involving complex sample matrices like stool, is often compromised by the presence of potent PCR inhibitors. These substances—including bile salts from the gastrointestinal tract, complex polysaccharides from dietary fiber, and humic acids from environmental contamination—can severely impair diagnostic accuracy and research outcomes in automated nucleic acid extraction workflows [49]. Inhibition mechanisms vary considerably, ranging from direct interference with DNA polymerase activity to disruption of fluorescence detection in real-time PCR [50]. Within the specific context of stool sample analysis, these inhibitors present a formidable barrier to reliable automated nucleic acid extraction and downstream molecular applications. This application note provides a detailed examination of the inhibition mechanisms of these key compounds and presents optimized, practical protocols to overcome their effects, ensuring successful genetic analysis in research and diagnostic settings.
Mechanisms of PCR Inhibition
PCR inhibitors present in challenging samples like stool operate through distinct biochemical mechanisms that disrupt the amplification process. Understanding these modes of action is critical for developing effective countermeasures.
Bile Salts: Bile acids and their salts, prevalent in gastrointestinal samples, directly inhibit the catalytic activity of DNA polymerases. Research has demonstrated significant variation in sensitivity to bile inhibition among different polymerase enzymes, with some like rTth showing relatively lower sensitivity compared to others like Taq [51]. The inhibitory effect is primarily attributed to the fraction containing bile acids and their salts, rather than bile proteins [51].
Complex Polysaccharides: These compounds, often originating from undigested plant material in the diet, act as potent PCR inhibitors in fecal samples [52] [53]. They are thought to interfere with the PCR reaction by mimicking DNA structure, potentially binding to DNA polymerases or other reaction components and thereby preventing efficient amplification of target sequences [53].
Humic Substances: As prominent inhibitors in environmental and soil-contaminated samples, humic acids and fulvic acids employ dual inhibition mechanisms. They not only suppress amplification by interacting with the DNA polymerase and nucleic acids but also act as potent detection inhibitors in real-time PCR [49] [50]. Humic acid specifically quenches the fluorescence of common double-stranded DNA binding dyes like SYBR Green I and EvaGreen through static or collisional quenching, where the humic acid molecules bind directly to the dye, leading to inaccurate quantification even when amplicon production remains unaffected [50].
Table 1: Characteristics of Major PCR Inhibitors in Complex Samples
| Inhibitor Class | Primary Sources | Mechanism of Inhibition | Impact on PCR |
|---|---|---|---|
| Bile Salts | Gastrointestinal samples, bile | Inhibition of polymerase catalytic activity [51] | Reduced amplification efficiency |
| Complex Polysaccharides | Dietary fiber, plant material, feces [52] [53] | Binding to polymerase; interference with nucleic acid interactions [53] | Complete or partial amplification failure |
| Humic Acids | Soil, sediment, environmental samples [49] | Polymerase interaction + fluorescence quenching of dsDNA dyes [50] | Suppressed amplification and inaccurate quantification |
Experimental Strategies and Protocols
Sample Preparation and Automated Nucleic Acid Extraction
Effective management of PCR inhibitors begins at the sample preparation stage, prior to automated nucleic acid extraction. For stool samples, mechanical lysis methods like bead-beating provide incremental yield improvements by effectively disrupting microbial cell walls, particularly for Gram-positive bacteria [54]. When selecting automated extraction systems, performance varies significantly between platforms in handling inhibitory substances. Comparative studies have revealed differences in yield, inter-sample variability, and subsequent sequencing readouts across commercial systems like the Bioer GenePure Pro, Promega Maxwell RSC 16, and ThermoFisher KingFisher Apex [54].
Specialized kits designed for inhibitor-rich samples incorporate specific technologies to overcome these challenges. For instance, the EZ2 PowerFecal Pro DNA/RNA Kit employs a patent-pending Inhibitor Removal Technology (IRT) and includes a crucial phenol chloroform isoamyl alcohol step for depletion of downstream inhibitors, enabling isolation of highly pure nucleic acids suitable for sensitive downstream applications like qPCR, dPCR, and next-generation sequencing [12]. The recommended starting material is 50-100 mg of stool, with stabilization options available for samples that cannot be processed immediately [12].
Optimization of PCR Components
Strategic selection and formulation of PCR reagents can substantially improve resistance to inhibitors:
DNA Polymerase Selection: Significant differences in inhibitor tolerance exist among DNA polymerases. For bile inhibition, rTth DNA polymerase has demonstrated lower sensitivity compared to Taq and Tfl enzymes [51]. Similarly, studies evaluating inhibitor-resistant PCR reagents for direct detection of pathogens in complex matrices found that no single chemistry performed best across all matrices, but Phusion Blood Direct PCR Kit, Phire Hot Start DNA polymerase, and KAPA Blood PCR kit showed particular promise for various challenging samples [55].
PCR Additives and Enhancers: Specific additives can counteract different inhibitor classes:
- Casein and Formamide: Effective against bile salts; adding casein (0.01% w/v) and formamide (1-5%) to the reaction mixture significantly reduces the PCR inhibitory effect of bile [51].
- BSA (Bovine Serum Albumin): Particularly useful for inhibitors including bile salts, feces, humic acid, and hemoglobin, typically used at concentrations of 0.01-0.1 μg/μl [56].
- Tween 20: Neutralizes inhibitors in feces and phenolic compounds; generally used at 0.5-2.5% concentration [56].
Table 2: PCR Additives for Specific Inhibitor Types
| Additive | Effective Against | Recommended Concentration | Mechanism of Action |
|---|---|---|---|
| Casein | Bile salts [51] | 0.01% (w/v) [56] | Binds to inhibitory compounds |
| Formamide | Bile salts, GC-rich templates [51] [56] | 1-5% [56] | Increases stringency; relieves inhibition |
| BSA | Bile salts, feces, humic acid, hemoglobin [56] | 0.01-0.1 μg/μl [56] | Binds inhibitors; stabilizes polymerase |
| Tween 20 | Feces, phenolic compounds, plant polysaccharides [56] | 0.5-2.5% [56] | Surfactant action; neutralizes SDS |
| Betaine | Hemoglobin, GC-rich templates [56] | 0.1-3.5M [56] | Reduces secondary structure; relieves inhibition |
Specialized Pre-PCR Treatment Protocols
Protocol for Bile-Rich Samples:
- Dilution and Heating: Dilute bile samples 1:10-1:100 in molecular grade water and heat at 98°C for 10 minutes [51].
- PCR Formulation: Prepare PCR mixture using rTth DNA polymerase supplemented with casein (0.01% w/v) and formamide (1-5% v/v) [51].
- Amplification: Perform amplification with an initial denaturation at 95°C for 5 minutes, followed by 40 cycles of denaturation at 95°C for 30 seconds, annealing at 55-60°C for 30 seconds, and extension at 72°C for 45 seconds [51].
Protocol for Humic Acid-Rich Environmental Samples:
- Nucleic Acid Extraction: Employ specialized purification methods with inhibitor removal technology, such as magnetic bead-based systems with multiple wash steps [12].
- Polymerase Selection: Use inhibitor-resistant polymerase blends specifically formulated for environmental samples [49] [55].
- Detection Chemistry: For quantitative PCR, consider switching to hydrolysis probe-based detection (e.g., TaqMan) instead of DNA-binding dyes, as hydrolysis probe fluorescence is not quenched by humic acid [50].
- Additive Supplementation: Include BSA (0.1 μg/μl) and Tween 20 (0.5-1%) in the PCR reaction to neutralize residual inhibitors [56].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for Overcoming PCR Inhibition
| Reagent/Category | Specific Examples | Function/Application |
|---|---|---|
| Inhibitor-Tolerant Polymerases | Phusion Blood Direct PCR Kit, Phire Hot Start DNA Polymerase, rTth DNA polymerase [51] [55] | Enhanced resistance to various inhibitors in complex samples |
| PCR Additives | BSA, Casein, Formamide, Tween 20, Betaine [51] [56] | Neutralize specific inhibitors; improve reaction efficiency |
| Specialized Buffers | STRboost, Ampdirect, PCRboost [55] | Chemical neutralization of inhibitory substances |
| Automated Extraction Systems | MagNA Pure Compact, KingFisher Flex, NucliSENS easyMAG [7] | High-throughput nucleic acid purification with inhibitor removal |
| Inhibitor-Removal Kits | EZ2 PowerFecal Pro DNA/RNA Kit with Inhibitor Removal Technology [12] | Specific removal of PCR inhibitors from challenging samples |
Workflow Diagram for Inhibitor Management
Figure 1: Strategic workflow for managing PCR inhibitors in automated nucleic acid analysis. This diagram outlines a systematic approach from sample collection through final detection, highlighting critical decision points for addressing different inhibitor types through specialized protocols.
Successfully overcoming PCR inhibitors—particularly bile salts, complex polysaccharides, and humic acids—in automated nucleic acid extraction from stool samples requires a multifaceted strategy that begins with appropriate sample handling and extends through optimized detection chemistry. The most effective approach combines specialized extraction methods with inhibitor-tolerant biochemical components, tailored to the specific inhibitory profile of the sample matrix. As molecular diagnostics continues to advance toward more automated and direct amplification methods, the implementation of these detailed protocols will be essential for obtaining reliable, reproducible results in both research and clinical settings.
Within the framework of automated nucleic acid extraction (ANE) research, consistent and reliable results from complex biological samples like stool are paramount. This application note details the critical pre-analytical steps of homogenization and clarification that are fundamental to successful downstream automated nucleic acid extraction and analysis. Stool samples present unique challenges due to their heterogeneous composition, varying consistency, and the presence of PCR inhibitors and undigested materials [57] [58]. Proper pre-processing mitigates these issues, ensuring that the nucleic acids delivered to the automated extraction system are representative of the entire sample and free from substances that could compromise analytical integrity. This protocol is designed for researchers and scientists developing and validating automated workflows for diagnostic, therapeutic, or genomic surveillance applications.
The Impact of Pre-processing on Downstream Analysis
The method of sample preparation prior to nucleic acid extraction significantly influences the yield, purity, and representativeness of the final genetic material. Inconsistent homogenization can lead to biased microbial community profiles, as different layers of a stool sample may contain varying microbial populations [58]. Furthermore, inadequate clarification can allow inhibitory substances to co-purify with nucleic acids, reducing the efficiency of downstream enzymatic reactions like PCR and sequencing [59] [57].
The choice of homogenization and disruption method is particularly critical for the detection of certain pathogens. For example, bead-beating, a harsh mechanical disruption technique, has been shown to be essential for the effective lysis of tough-walled helminth eggs and Gram-positive bacteria. One study demonstrated that omitting bead-beating led to a 5-fold decrease in the detection of Blautia and a 14-fold decrease in Bifidobacterium, significantly altering the perceived microbial composition [58]. The table below summarizes key findings from the literature on the effects of pre-processing variables.
Table 1: Effects of Pre-processing Variables on Nucleic Acid Analysis from Stool Samples
| Variable | Effect on Analysis | Key Evidence |
|---|---|---|
| Incomplete Homogenization | Altered microbial community profile; under-representation of specific taxa. | Minor differences in species abundance between inner and outer stool layers; can bias individual sample profiles [58]. |
| Omission of Bead-Beating | Significant under-detection of Gram-positive bacteria and helminths. | Proportions of Blautia and Bifidobacterium decreased by 5- and 14-fold, respectively; poor recovery of helminth DNA in 20% of PCR assays [57] [58]. |
| Sample Dilution (Simulated Diarrhea) | Increased genomic DNA fragmentation upon freezing. | No significant effect on 16S rRNA gene sequencing profile; may impact shotgun metagenomics [58]. |
| Clarification Efficiency | Reduction of PCR inhibitors and non-target debris. | Critical for maximizing sensitivity of downstream real-time PCR and NGS; prevents column clogging in automated systems [60] [61]. |
Materials and Reagents
Research Reagent Solutions
The following table lists essential materials and their functions for the pre-processing of stool samples.
Table 2: Essential Reagents and Materials for Stool Sample Pre-processing
| Item | Function/Application |
|---|---|
| Phosphate-Buffered Saline (PBS) | A neutral buffer for suspending and diluting stool samples without damaging microbial cells [58]. |
| Formalin (10%) | A preservative for fixed stool specimens; allows for concentration procedures for parasitic organisms [60]. |
| Polyvinyl Alcohol (PVA) | A preservative for stool samples intended for permanent staining and microscopic examination [60]. |
| SAF (Sodium Acetate-Acetic Acid-Formalin) Solution | A preservative and enrichment solution used for the microscopic detection of parasites [57]. |
| Ethyl Acetate | A solvent used in the formalin-ethyl acetate sedimentation concentration technique to separate fat and debris from parasites [60]. |
| Silica-coated Magnetic Beads | The solid phase for binding nucleic acids in many automated extraction systems; integrated into automated liquid handling protocols [62] [63]. |
| Lysis Buffer (with Chaotropic Salts) | Disrupts cells and virions, denatures proteins, and creates conditions for nucleic acid binding to silica surfaces [59] [63]. |
| Proteinase K | An enzyme that digests proteins and enhances the lysis of tough microbial structures, often used in conjunction with bead-beating [57]. |
| DNase/RNase-free Beads | Inert beads for mechanical disruption of microbial cell walls in a bead beater [57] [58]. |
| Transport Medium (e.g., MEM) | Maintains viral viability and integrity in samples destined for viral metagenomics or culture [61]. |
Experimental Protocols
Protocol 1: Standard Homogenization and Clarification for Automated ANE
This protocol is designed for the pre-processing of fresh or frozen stool samples prior to automated nucleic acid extraction, optimized for bacterial and viral metagenomics or pathogen detection.
Materials:
- Sterile PBS
- 2 mL screw-cap tubes with O-rings
- DNase/RNase-free bead-beating tubes (e.g., containing 0.1mm glass or ceramic beads)
- Low-speed centrifuge
- Vortex mixer
- Bead beater
Procedure:
- Weighing: Accurately weigh 100-200 mg of stool into a 2 mL screw-cap tube.
- Suspension: Add a pre-determined volume of sterile PBS to create a 10-20% (w/v) suspension. For example, add 900 µL of PBS to 100 mg of stool.
- Homogenization: Vortex the mixture vigorously for at least 1 minute or until the stool is fully suspended and no large particulate matter remains.
- Clarification (Low-Speed Centrifugation):
- Centrifuge the homogenate at 500 × g for 5 minutes at 4°C. This low-speed step pellets undigested food particles, eukaryotic cells, and large debris while leaving bacteria, viruses, and soluble factors in the supernatant [61].
- Aliquoting for Extraction: Carefully transfer the clarified supernatant to a new, sterile tube. This supernatant is now ready to be used as the input for your automated nucleic acid extraction system.
Protocol 2: Harsh Lysis with Bead-Beating for Helminth and Gram-Positive Bacteria Detection
This method is critical for the efficient recovery of nucleic acids from organisms with robust cell walls or cysts, such as helminth eggs or Gram-positive bacteria [57].
Materials:
- Lysis buffer (e.g., from QIAamp DNA Stool Mini Kit or equivalent)
- Proteinase K
- Bead beater and DNase/RNase-free bead-beating tubes
Procedure:
- Initial Processing: Follow steps 1-3 of Protocol 1 to create a homogenized stool suspension in PBS.
- Lysis: Transfer 200 µL of the homogenate to a bead-beating tube. Add the recommended volumes of lysis buffer and Proteinase K according to your chosen extraction kit.
- Harsh Mechanical Disruption:
- Incubate the mixture as per the kit protocol (typically at 56°C for 30 minutes) to enable enzymatic digestion.
- Secure the tubes in a bead beater and process at high speed for 1-3 minutes.
- Clarification: Centrifuge the lysate at high speed (e.g., >10,000 × g for 1 minute) to pellet beads and insoluble debris.
- Aliquoting for Extraction: The resulting supernatant contains the released nucleic acids and is suitable for automated extraction. Note: Some automated systems can integrate the clarified lysate directly, while others may require an additional centrifugation step if the sample is not fully clear.
Protocol 3: Formalin-Ethyl Acetate Sedimentation for Parasite Concentration
This CDC-recommended procedure is a standard for concentrating parasitic organisms from preserved stool samples and can be adapted to provide input for nucleic acid extraction [60].
Materials:
- 10% Formalin
- Ethyl Acetate
- 15 mL conical centrifuge tubes
- Cheesecloth or gauze
- Saline (0.85%) or 10% formalin
Procedure:
- Preservation: The stool sample should be preserved in 10% formalin or PVA.
- Straining: Mix the specimen well. Strain approximately 5 mL of the fecal suspension through wetted gauze into a 15 mL conical centrifuge tube.
- Washing: Add saline or 10% formalin through the debris on the gauze to bring the volume to 15 mL.
- First Centrifugation: Centrifuge at 500 × g for 10 minutes. Decant the supernatant.
- Formalin and Ethyl Acetate Addition: Resuspend the sediment in 10 mL of 10% formalin. Add 4 mL of ethyl acetate, stopper the tube, and shake vigorously for 30 seconds.
- Second Centrifugation: Centrifuge again at 500 × g for 10 minutes. Four layers will form: ethyl acetate, plug of debris, formalin, and sediment.
- Sediment Collection: Free the debris plug with an applicator stick and decant the top three layers. The purified sediment at the bottom contains the concentrated parasites and can be used for nucleic acid extraction or microscopy.
Workflow Visualization
The following diagram illustrates the logical workflow for selecting the appropriate pre-processing path based on research objectives.
Discussion
The pre-analytical phase is a significant source of variability in microbiome and pathogen detection studies. The protocols detailed here provide a standardized approach to homogenization and clarification, which are critical for generating reproducible and accurate data in automated nucleic acid extraction workflows from stool samples. The choice of protocol must be driven by the specific research question, as the optimal method for detecting labile viruses is fundamentally different from that required for robust helminth eggs [57] [61]. Integrating these validated pre-processing steps with automated ANE systems enhances throughput, reduces human error, and ensures that the input material is of consistent quality, thereby unlocking the full potential of downstream high-resolution molecular analyses [62] [63].
Automated nucleic acid extraction from stool samples presents significant challenges for researchers and drug development professionals due to the complex nature of the sample matrix. Stool contains potent PCR inhibitors and a diverse mixture of microorganisms with varying cell wall structures, making optimization of each extraction parameter critical for obtaining reliable results. This application note provides a detailed framework for optimizing three fundamental parameters—lysis conditions, binding efficiency, and wash stringency—to maximize nucleic acid yield, purity, and reproducibility in automated extraction systems. The protocols outlined here are essential for applications in microbiome research, pathogen detection, and clinical diagnostics where stool serves as a primary sample matrix.
Lysis Conditions Optimization
Effective cell lysis is the first critical step in nucleic acid extraction from stool samples. The complex composition of stool, containing both Gram-positive and Gram-negative bacteria with differing cell wall structures, necessitates optimized lysis conditions for comprehensive nucleic acid recovery.
Lysis Method Comparison
Table 1: Comparison of Lysis Techniques for Stool Samples
| Lysis Method | Mechanism | Application | Efficiency on Gram-positive Bacteria | Integration with Automated Platforms |
|---|---|---|---|---|
| Bead-beating | Mechanical shearing | Total community DNA extraction | High | Requires pre-lysis module |
| Chemical Detergents | Membrane disruption | Soluble protein extraction | Moderate | Compatible |
| Laser Pulse | Shock wave generation | Single-cell analysis | High | Limited compatibility |
| Electrical | Electroporation | Rapid lysis for enzymes | Variable | Moderate compatibility |
Recent comparative studies demonstrate that bead-beating provides incremental yield improvements by effectively lysing difficult-to-disrupt organisms compared to lysis buffer alone [54]. This method is particularly crucial for the representation of Gram-positive bacteria in human fecal microbiota research, as their thick peptidoglycan layer resists standard chemical lysis methods [54].
Automated Platform Lysis Performance
Table 2: Lysis Efficiency of Automated Nucleic Acid Extraction Systems for Stool
| Extraction Platform | Lysis Method | Inhibitor Removal Efficiency | qRT-PCR Consistency | Conventional RT-PCR Performance |
|---|---|---|---|---|
| MagNA Pure Compact | Not specified | High | Most consistent | Positive in all cases |
| KingFisher Flex | Not specified | Moderate | Good | Positive in all cases |
| NucliSENS easyMAG | Not specified | Moderate | Variable | Not detected in all cases |
| QIAamp Viral RNA kit | Manual | Lower (inhibitors detected) | Variable | Inhibitors detected |
Evaluation of six extraction methods for rotavirus RNA detection from stool samples demonstrated that extracts prepared using the MagNA Pure Compact instrument yielded the most consistent results by both qRT-PCR and conventional RT-PCR [7]. This platform effectively removed RT-PCR inhibitors known to be present in stool samples, a common challenge in stool-based nucleic acid extraction.
Experimental Protocol: Lysis Optimization for Stool Samples
Materials:
- Stool samples (aliquoted and stored at -80°C)
- Lysis buffers: NP-40, RIPA, or commercial formulations
- Bead-beating system with different bead sizes
- Proteinase K
- Thermal shaker
- Automated extraction platforms (e.g., MagNA Pure Compact, KingFisher Flex)
Method:
- Sample Preparation: Homogenize stool samples in appropriate buffer (e.g., PBS) to create 10% suspensions. Centrifuge briefly to remove large particulates.
- Lysis Condition Testing: Divide each sample into aliquots for different lysis treatments:
- Chemical Lysis Only: Incubate with lysis buffer (NP-40 or RIPA) containing 1 mg/mL Proteinase K at 56°C for 30 minutes.
- Mechanical Lysis: Process samples using bead-beating with 0.1mm glass beads for 30-90 seconds.
- Combined Approach: Perform bead-beating followed by chemical lysis.
- Nucleic Acid Extraction: Process lysates through automated extraction platforms using consistent binding and wash steps.
- Downstream Analysis: Quantify nucleic acid yield and purity using spectrophotometry. Assess quality via 16S rRNA gene amplification (for bacterial community analysis) or pathogen-specific PCR.
Optimization Parameters:
- Bead-beating duration: Test 30, 60, 90-second intervals
- Lysis buffer composition: Vary detergent concentrations (0.1-2% for nonionic detergents)
- Enzymatic treatment: Optimize Proteinase K concentration and incubation time
Binding Efficiency Optimization
Binding efficiency determines the recovery of nucleic acids from the lysate. For stool samples, this step must be optimized to overcome inhibitors and compete with non-specific binding to sample components.
Binding Buffer Composition Optimization
Research with polyethyleneimine-coated iron oxide nanoparticles (PEI-IONPs) demonstrates that binding buffer composition significantly impacts DNA adsorption efficiency [64]. Systematic optimization of key components yielded dramatically different results:
Table 3: Effect of Binding Buffer Composition on DNA Recovery Using PEI-IONPs
| PEG-6000 Concentration | NaCl Concentration | pH | DNA Concentration (ng/μL) | DNA Yield (μg) | Purity (A260/A280) |
|---|---|---|---|---|---|
| 30% | 0M | 4.0 | 34 ± 1.2 | 6.8 ± 0.2 | 1.81 |
| 10% | 0.5M | 7.0 | 18 ± 0.8 | 3.6 ± 0.1 | 1.65 |
| 20% | 0.25M | 5.5 | 25 ± 1.0 | 5.0 ± 0.2 | 1.72 |
The optimal binding buffer composition consisted of PEG-6000 concentration of 30%, NaCl concentration of 0M, and pH of 4, which yielded the highest DNA concentration, yield, and purity [64]. The low ionic strength minimizes the shielding effect, allowing strong electrostatic interactions between the negatively charged DNA backbone and positively charged binding surfaces.
Mechanism of Binding Enhancement
The binding mechanism involves several synergistic effects:
- PEG creates a crowded macromolecular environment that increases the effective concentration of DNA molecules, promoting aggregation and precipitation through osmotic pressure that reduces DNA solubility [64].
- Low NaCl concentration minimizes charge shielding, allowing strong electrostatic interactions between the negatively charged DNA phosphate backbone and positively charged binding surfaces [64].
- Acidic pH (4.0) may enhance binding by modulating the charge states of both the nucleic acids and binding surfaces.
Experimental Protocol: Binding Efficiency Optimization
Materials:
- Polyethyleneimine-coated iron oxide nanoparticles (PEI-IONPs)
- Binding buffers with varying PEG concentrations (10%, 20%, 30%)
- NaCl solutions (0M, 0.25M, 0.5M)
- pH-adjusted buffers (4.0, 5.5, 7.0)
- Magnetic separation rack
- UV-Vis spectrophotometer for quantification
Method:
- Nanoparticle Preparation: Synthesize PEI-IONPs using co-precipitation method and characterize for size and surface charge [64].
- Buffer Preparation: Prepare binding buffers with systematic variation in PEG-6000 concentration (10-30%), NaCl concentration (0-0.5M), and pH (4.0-7.0).
- Binding Reaction: Mix constant amount of stool lysate with nanoparticles in different binding buffers. Incubate with rotation for 15-30 minutes at room temperature.
- Magnetic Separation: Place tubes on magnetic rack for 2-5 minutes until solution clears. Carefully remove and retain supernatant.
- Efficiency Calculation: Quantify DNA in both bound (after elution) and unbound (supernatant) fractions to calculate binding efficiency:
- Binding Efficiency (%) = (Bound DNA / Total DNA) × 100
- Downstream Application: Test optimized binding conditions in PCR amplification of 16S rRNA genes and pathogen-specific targets.
Optimization Parameters:
- Incubation time: Test 5, 15, 30-minute intervals
- PEG molecular weight: Compare PEG-4000, PEG-6000, PEG-8000
- Binding capacity: Determine maximum nucleic acid binding per mg nanoparticles
Wash Stringency Optimization
Wash steps are critical for removing PCR inhibitors and non-specifically bound molecules without eluting the target nucleic acids. In stool samples, this balance is particularly challenging due to the high inhibitor content.
Principles of Wash Stringency
Stringency in wash buffers determines the specificity of nucleic acid retention during purification. The fundamental parameters controlling stringency are temperature and salt concentration [65]:
- High stringency conditions favor detection of only perfectly matched hybrids by disrupting imperfect binding
- Low stringency conditions preserve both specific and non-specific interactions
Automated Platform Wash Performance
Comparative studies of automated extraction systems reveal significant differences in inhibitor removal efficiency during wash steps:
Table 4: Wash Efficiency of Automated Nucleic Acid Extraction Platforms for Stool Samples
| Extraction Platform | Wash Efficiency | Inhibitor Removal | Nucleic Acid Retention | Recommended Applications |
|---|---|---|---|---|
| MagNA Pure Compact | High | Effective | High | Sensitive detection, genotyping |
| KingFisher Flex | High | Effective | High | Routine diagnostics |
| NucliSENS easyMAG | Moderate | Variable | Moderate | High-throughput screening |
| QIAamp Viral RNA kit | Lower | Inefficient (inhibitors detected) | High | Clean sample types |
The MagNA Pure Compact and KingFisher Flex systems demonstrated superior wash efficiency, with rotavirus RNA detected in all samples by conventional RT-PCR, while other methods showed variable performance [7].
Experimental Protocol: Wash Stringency Optimization
Materials:
- Automated nucleic acid extraction platforms
- Wash buffers with varying salt concentrations (0.1X SSC to 2X SSC)
- Temperature-controlled incubation blocks
- PCR reagents for inhibitor detection
Method:
- Platform Selection: Choose automated extraction systems with customizable wash protocols.
- Wash Buffer Preparation: Prepare wash buffers with varying salt concentrations:
- Low stringency: 2X SSC (standard saline citrate)
- Medium stringency: 1X SSC
- High stringency: 0.1X SSC
- Temperature Optimization: For each salt concentration, test wash temperatures ranging from 25°C to 65°C.
- Wash Protocol Implementation: Program automated platforms to use the optimized wash conditions:
- Number of wash cycles (typically 2-5)
- Wash volume (200-300 μL as recommended for ELISA protocols) [66]
- Incubation time for each wash (30 seconds to 2 minutes)
- Efficiency Assessment: Evaluate wash efficiency by:
- Inhibitor Detection: Spike samples with internal control DNA and measure PCR efficiency
- Purity Measurement: A260/A280 and A260/A230 ratios
- Yield Comparison: Total nucleic acid recovery after washes
Optimization Parameters:
- Salt concentration: Test 0.1X, 0.5X, 1X, 2X SSC
- Wash temperature: 25°C, 37°C, 45°C, 55°C, 65°C
- Wash volume: 200μL, 300μL, custom volumes based on platform
- Number of washes: 2, 3, 4, 5 cycles
Integrated Workflow and The Scientist's Toolkit
Successful nucleic acid extraction from stool samples requires integration of all optimized parameters into a cohesive workflow. The following toolkit summarizes essential reagents and their functions for implementing these optimized protocols.
Research Reagent Solutions
Table 5: Essential Reagents for Automated Nucleic Acid Extraction from Stool
| Reagent Category | Specific Examples | Function | Optimization Tips |
|---|---|---|---|
| Lysis Reagents | NP-40, RIPA buffer, Proteinase K, bead-beating matrices | Cell disruption and nucleic acid release | Combine mechanical and chemical methods for Gram-positive bacteria |
| Binding Enhancers | PEG-6000, PEI-IONPs, silica-coated magnetic particles | Nucleic acid immobilization | Use 30% PEG-6000, 0M NaCl, pH 4.0 with PEI-IONPs |
| Wash Buffers | SSC buffer (varying concentrations), ethanol-based washes | Contaminant and inhibitor removal | Higher temperature + lower salt increases stringency |
| Elution Buffers | TE buffer, nuclease-free water | Nucleic acid recovery | Pre-warm to 65°C for higher yields |
| Inhibition Controls | Internal control DNA, PCR efficiency markers | Process quality assessment | Include in every extraction batch |
Integrated Optimization Workflow
Optimization of lysis conditions, binding efficiency, and wash stringency parameters significantly impacts the success of automated nucleic acid extraction from challenging stool samples. Integration of mechanical lysis through bead-beating with chemical lysis ensures comprehensive disruption of diverse microbial communities. Employment of optimized binding buffers with 30% PEG-6000, 0M NaCl, and pH 4.0 maximizes nucleic acid recovery through enhanced electrostatic interactions. Implementation of high-stringency wash conditions through elevated temperature and reduced salt concentration effectively removes PCR inhibitors while maintaining target nucleic acid binding. These optimized parameters collectively address the specific challenges of stool samples, enabling reliable, reproducible nucleic acid extraction for sensitive downstream applications in research and diagnostic contexts.
Within the context of automated nucleic acid extraction from stool samples, rigorous RNA quality control (QC) is not merely a preliminary step but a critical determinant of research success. The complex composition of stool, rich in PCR inhibitors and diverse microbial communities, presents unique challenges for obtaining high-quality RNA suitable for sensitive downstream applications like RT-PCR, qPCR, and next-generation sequencing (NGS) [12] [5]. This application note provides a detailed framework for assessing RNA purity, integrity, and concentration, delivering specific protocols and metrics to ensure the reliability of data derived from automated extraction systems.
RNA Quantity and Purity Assessment
Accurate quantification and purity evaluation are the first critical steps post-extraction, as impurities can severely inhibit enzymatic reactions in downstream assays [67].
Spectrophotometric Analysis (UV-Vis)
Spectrophotometry measures the absorption of ultraviolet light by nucleic acids at 260 nm, providing a rapid, non-destructive method for quantifying RNA concentration and assessing purity through absorbance ratios [67] [68].
Protocol: Using a Microvolume Spectrophotometer
- Blank Instrument: Use the same elution buffer (e.g., RNase-free water or TE buffer) used for your RNA sample.
- Load Sample: Pipette 1-2 µL of the RNA sample onto the measurement pedestal.
- Measure and Record: Lower the arm and initiate the measurement. Record the following:
- Concentration: Calculated from the A260 reading. An A260 of 1.0 is equivalent to ~40 µg/mL for RNA [69].
- Purity Ratios: A260/A280 and A260/A230.
- Interpret Results: Compare the calculated ratios against the accepted thresholds for pure RNA (Table 1).
Table 1: Interpretation of Spectrophotometric RNA Quality Metrics
| Metric | Target Value for Pure RNA | Significance of Deviation |
|---|---|---|
| A260/A280 Ratio | ~2.0 (1.8-2.1 is acceptable) [67] | A lower ratio suggests protein contamination (e.g., from incomplete lysis) [67] [68]. |
| A260/A230 Ratio | >1.8 [67] [69] | A lower ratio indicates contamination by salts, organic compounds, or chaotropic agents (e.g., guanidine) carried over from the extraction kit [67] [69]. |
| Concentration | Application-dependent | Overestimation can occur from DNA contamination or residual ethanol [68]. |
Fluorometric Analysis
For samples with low yields, such as those from low-biomass stool samples, fluorometry provides superior sensitivity and specificity over spectrophotometry [67] [70].
Protocol: Using RNA-Specific Fluorescent Dyes (e.g., RiboGreen)
- Prepare Standards: Create a dilution series of a known-concentration RNA standard in the same buffer as your samples.
- Prepare Dye Working Solution: Dilute the fluorescent dye according to the manufacturer's instructions.
- Mix Samples and Standards: Combine equal volumes of each standard/unknown sample with the dye working solution in a microplate or PCR tubes. Incubate for a brief period (e.g., 2-5 minutes) protected from light.
- Measure Fluorescence: Use a fluorometer or microplate reader to measure the fluorescence at the appropriate excitation/emission wavelengths.
- Generate Standard Curve and Calculate Concentration: Plot the fluorescence of the standards against their known concentrations. Use the resulting curve's linear regression equation to calculate the concentration of the unknown samples [69] [70].
RNA Integrity Assessment
RNA integrity is paramount for applications requiring full-length transcripts. Degradation, a major concern due to ubiquitous RNases, can be systematically evaluated.
Automated Capillary Electrophoresis
This is the gold standard for objectively evaluating RNA integrity, providing an RNA Integrity Number (RIN) or similar score [67] [70].
Protocol: Using an Agilent 2100 Bioanalyzer
- Prepare the Chip: Prime the specific RNA chip (e.g., RNA 6000 Nano) with the provided gel-dye mix.
- Prepare RNA Ladder and Samples: Mix the RNA ladder and your RNA samples (typically 1 µL at 10-500 ng/µL) with specific markers and denaturing agents as per the kit instructions.
- Load the Chip: Pipette the ladder and samples into the designated wells.
- Run and Analyze: Place the chip in the instrument and start the run. The software will generate an electropherogram and a gel-like image, automatically calculating the concentration and RIN score [71] [70].
Table 2: Interpretation of RNA Integrity Metrics from Capillary Electrophoresis
| Metric | Ideal Result | Interpretation |
|---|---|---|
| RNA Integrity Number (RIN) | 10 (perfectly intact) [70] | A score >8 is considered perfect for downstream applications. A score >5 is often the minimum acceptable threshold for qRT-PCR [72]. |
| 28S:18S rRNA Ratio | ~2:1 (for eukaryotic samples) [71] | A decreased ratio indicates degradation. Note: This metric is less reliable for bacterial RNA or RNA from FFPE samples [69]. |
| Electropherogram Profile | Sharp ribosomal peaks with a flat baseline and minimal low-molecular-weight smear [71] | A smeared profile or shift to shorter retention times indicates widespread degradation [71] [73]. |
Agarose Gel Electrophoresis (Denaturing)
A traditional, cost-effective method for visually assessing RNA integrity, though it requires more RNA and is less quantitative [71] [74].
Protocol: "Bleach Gel" Electrophoresis This protocol offers a safe, simple, and effective denaturing gel method by incorporating commercial bleach into a standard TAE agarose gel [74].
- Prepare Gel Solution: For a 1% gel, mix 0.5 g agarose with 50 mL of 1x TAE buffer.
- Add Bleach: Add 250-500 µL of commercial bleach (6% sodium hypochlorite) per 50 mL of gel solution to achieve a 0.5-1% v/v final concentration. Note: Add bleach BEFORE melting the agarose.
- Melt and Cool: Heat the mixture in a microwave to melt the agarose, then cool to ~60°C.
- Add Stain and Pour: Add an intercalating dye like ethidium bromide or a safer alternative (e.g., SYBR Gold), then pour the gel and allow it to solidify [74].
- Prepare and Load Samples: Mix RNA samples (100-200 ng recommended) with a standard DNA loading buffer.
- Run Gel: Submerge the gel in 1x TAE buffer and run at constant voltage (e.g., 100 V) for ~35 minutes.
- Visualize: Image the gel under UV transillumination. Intact eukaryotic RNA will display sharp 28S and 18S rRNA bands with a 2:1 intensity ratio. Degraded RNA will appear as a smeared smear without distinct bands [71] [74].
The Scientist's Toolkit: Essential Reagents for RNA QC
Table 3: Key Research Reagent Solutions for RNA Quality Control
| Reagent / Kit | Function / Application |
|---|---|
| DNase I (RNase-free) | Digests contaminating genomic DNA in RNA samples to ensure accurate quantification and prevent false positives in RT-PCR [67] [68]. |
| RNA-Specific Fluorescent Dyes (e.g., RiboGreen) | Highly sensitive and specific quantification of RNA concentration, especially for low-yield samples [70] [68]. |
| Agilent RNA 6000 Nano/Pico Kit | Microfluidics-based kit for use with the Bioanalyzer system to assess RNA integrity, concentration, and generate a RIN score [71] [70]. |
| SYBR Gold / SYBR Green II | Highly sensitive fluorescent nucleic acid gels stains used as safer, more sensitive alternatives to ethidium bromide for gel electrophoresis [71] [69]. |
| Inhibitor Removal Technology (IRT) Kits | Specialized reagents in kits (e.g., QIAGEN EZ2 PowerFecal Pro) designed to co-purify and remove PCR inhibitors common in complex samples like stool [12]. |
Experimental Workflow and Data Interpretation
The following diagram illustrates the logical workflow for comprehensive RNA quality assessment, from sample preparation to decision-making for downstream applications.
Special Considerations for Stool Samples
Automated extraction of RNA from stool samples requires specific considerations to ensure quality.
- Inhibitor Removal: Stool contains complex PCR inhibitors. Use kits with dedicated Inhibitor Removal Technology (IRT), like the QIAGEN EZ2 PowerFecal Pro, to obtain highly pure nucleic acids [12].
- Mechanical Lysis: Efficient lysis of diverse microbial cells (including Gram-positive bacteria) is critical for representative RNA yields. Bead-beating is highly recommended and should be integrated prior to automated extraction [5].
- Sample Stabilization: To preserve RNA integrity, process stool samples immediately after collection or stabilize them using reagents like DNA/RNA Shield [12] [5].
The following workflow details the specialized process for handling stool samples:
A rigorous, multi-faceted QC pipeline is non-negotiable for research based on RNA from automated stool extraction. By systematically applying the quantification, purity, and integrity assessments outlined here—and interpreting them within the context of stool-specific challenges—researchers can confidently select high-quality RNA samples, thereby ensuring the robustness and reproducibility of their gene expression data in drug development and clinical research.
The reliable analysis of nucleic acids from low-biomass stool samples presents a significant challenge in molecular diagnostics and microbiome research. The low abundance of microbial cells, coupled with the presence of potent PCR inhibitors such as complex polysaccharides and bilirubin, can severely compromise extraction efficiency and downstream application results. Within the broader context of automated nucleic acid extraction research, achieving consistent, high-yield recovery from these challenging specimens is paramount for obtaining accurate and reproducible data. This application note details evidence-based techniques and optimized protocols to overcome these hurdles, ensuring the integrity of your research and drug development workflows.
Critical Factors Influencing Yield in Low-Biomass Samples
The efficiency of nucleic acid recovery from low-biomass samples is governed by several interconnected factors. Understanding these variables is the first step in optimizing any extraction protocol.
- Inhibitor Removal: Stool samples contain strong reverse transcription-polymerase chain reaction (RT-PCR) inhibitors that can persist through inadequate extraction methods, directly impacting detection sensitivity [7]. Effective extraction must robustly remove these contaminants.
- Bacterial Biomass and Lysis Efficiency: Specimen bacterial biomass is a key driver of 16S rRNA gene sequencing profiles [75]. In low-biomass contexts, the DNA extraction method and the efficiency with which it can lyse diverse microbial cell types (e.g., easy-to-lyse versus hard-to-lyse bacteria) become critically important for representative recovery [75].
- Contamination and Background Signal: Low endogenous DNA levels make results highly susceptible to contamination from reagents or the laboratory environment [75]. This can lead to high alpha diversities and spurious operational taxonomic units (OTUs) that do not represent the true sample composition.
- Technical Reproducibility: Low biomass technical repeats have been shown to produce reduced sequencing reproducibility, underscoring the need for optimized and consistent protocols [75].
Comparative Evaluation of Extraction Systems and Methodologies
Selecting the appropriate extraction platform and methodology is fundamental. The following table summarizes the performance of various systems as evaluated in comparative studies, providing a basis for informed selection.
Table 1: Performance Comparison of Nucleic Acid Extraction Methods for Stool and Low-Biomass Samples
| Extraction Method / Platform | Sample Type | Key Findings Related to Low-Biomass Yield | Study Reference |
|---|---|---|---|
| MagNA Pure Compact (Roche) | Stool (Rotavirus) | Yielded the most consistent results by qRT-PCR and conventional RT-PCR; detected rotavirus in all dilution series samples. | [7] |
| KingFisher Flex (Thermo Fisher) | Stool (Rotavirus) | Alongside MagNA Pure, successfully detected rotavirus in all dilution series samples by conventional RT-PCR. | [7] |
| NucliSENS easyMAG (bioMérieux) | Stool (Norovirus), Plasma (CMV) | One of five automated platforms that yielded comparable results for viral extraction, though performance could be impaired by inhibitors in stool. | [6] |
| QIAamp Viral RNA Mini Kit (Manual) | Stool (Rotavirus) | RT-PCR inhibitors were detected in extracts produced with this manual kit. | [7] |
| Kit-QS (DSP Virus/Pathogen Kit) | Bacterial Mock Communities | Better represented hard-to-lyse bacteria from mock communities compared to Kit-ZB; produced purer DNA. | [75] |
| Bead-Beating | Human Fecal Samples | Incremental yield increase; greater representation of Gram-positive bacteria; considered gold standard for effective lysis of different microbial cell types. | [5] |
The Role of Mechanical Lysis
A critical finding from comparative studies is the importance of mechanical lysis. Research on human fecal samples has demonstrated that bead-beating provides an incremental yield compared to using lysis buffer alone. This method is essential for effectively lysing a wide range of microbial cell types, including spores, and is particularly crucial for achieving a greater representation of Gram-positive bacteria, which have more robust cell walls [5]. The International Human Microbiome Standards project includes bead-beating in its standard operating procedures for fecal samples, solidifying its status as a best practice [5].
Optimized Protocol for Automated Extraction from Low-Biomass Stool
The following protocol is synthesized from best practices identified in the cited research, designed for integration with automated magnetic bead-based extraction systems like the KingFisher Apex or MagNA Pure Compact.
Materials and Reagents
Table 2: Research Reagent Solutions for Low-Biomass Stool Extraction
| Reagent / Kit | Function | Considerations for Low-Biomass |
|---|---|---|
| DNA/RNA Shield (e.g., from Zymo Research) | Preservation reagent; nuclease inhibition | Preserves nucleic acid integrity during storage, critical for maintaining already low target levels. |
| Lysis Buffer with Proteinase K | Cellular lysis and protein digestion | Essential for breaking down sample structure and degrading proteins. |
| MagMAX Microbiome Ultra Kit (Thermo Fisher) or equivalent | Magnetic bead-based purification | Optimized for microbiome studies; includes reagents for inhibitor removal. |
| Phenol-Chloroform-Isoamyl Alcohol | Organic extraction and protein removal | Highly effective for removing proteins and lipids; requires careful handling of toxic solvents [76]. |
| Bead Beating Tubes (Lysing Matrix E) | Mechanical disruption | Ensures lysis of hard-to-break microbial cells (e.g., Gram-positives, spores) [5]. |
| RNase A and/or DNase I | Removal of contaminating nucleic acids | For target-specific applications (e.g., DNA-only for 16S sequencing). |
| Ethanol/Isopropanol | Nucleic acid precipitation | Concentrates nucleic acids, improving yield from dilute extractions. |
Step-by-Step Workflow
Sample Preservation and Homogenization
- Collect stool sample immediately into an appropriate preservation reagent such as DNA/RNA Shield [5]. For frozen samples, avoid multiple freeze-thaw cycles.
- Thoroughly homogenize the sample by vortexing. For solid stools, this may require initial suspension in the preservation buffer.
Mechanical Lysis (Bead-Beating)
- Transfer a 300 µL aliquot of the homogenized sample to a tube containing a lysing matrix (e.g., garnet or silica beads).
- Process using a homogenizer like the FastPrep-24 at 6.0 m/s for 40 seconds [5]. This step is non-negotiable for comprehensive cell disruption.
Optional Organic Extraction for Challenging Samples
- For samples with persistent inhibitors, add an organic extraction step using phenol-chloroform-isoamyl alcohol (25:24:1) [76].
- Centrifuge to separate phases. The aqueous upper phase, containing the nucleic acids, should be carefully transferred to a new tube for the automated protocol.
Automated Nucleic Acid Purification
- Load the supernatant (or the aqueous phase from the organic extraction) onto the automated platform alongside the manufacturer's recommended magnetic beads and wash buffers.
- Use platforms with built-in UV sterilization (e.g., KingFisher Apex, GenePure Pro) to minimize cross-contamination [5].
- Elute in a small volume (e.g., 50-100 µL) of nuclease-free water or TE buffer to maximize final concentration.
Quality Control and Validation
- Quantification and Purity: Use a fluorometer (e.g., Qubit) for accurate nucleic acid quantification and a spectrophotometer (e.g., NanoDrop) to check for residual contaminants (A260/A280 and A260/A230 ratios) [5].
- Inhibition Testing: Spike extracts with a known quantity of exogenous control DNA/RNA and perform PCR to check for the presence of inhibitors [7].
- Process Controls: Include a known mock microbial community (e.g., ZymoBIOMICS Microbial Community Standard) and no-template controls (NTCs) in each extraction batch to monitor performance, contamination, and bias [75] [5].
The following workflow diagram illustrates the optimized protocol for automated extraction.
Optimizing recovery from low-biomass stool samples requires a multifaceted approach that prioritizes effective cell lysis, rigorous inhibitor removal, and contamination control. The integration of mandatory bead-beating for mechanical disruption and the strategic selection of automated extraction platforms proven to handle inhibitors—such as the MagNA Pure Compact or KingFisher systems—form the foundation of a robust protocol. By adhering to the detailed methodologies and quality control measures outlined in this application note, researchers and drug development professionals can significantly improve the yield, purity, and reliability of nucleic acids extracted from these challenging samples, thereby enhancing the validity of their downstream analyses.
Benchmarking Performance: Analytical and Clinical Validation Frameworks
In the development and validation of molecular assays, particularly for complex matrices like stool samples, establishing the Limit of Detection (LoD) is a critical analytical parameter. The LoD represents the lowest concentration of an analyte that can be reliably distinguished from a blank or negative sample [77]. For laboratory-developed tests (LDTs) and diagnostic assays, regulatory guidelines such as CLIA require clinical laboratories to establish and document this analytical sensitivity before patient implementation [78]. Probit analysis has emerged as a powerful statistical method for LoD determination, especially for qualitative and quantitative molecular assays that exhibit binary outcomes (positive/negative) [77]. This approach is particularly valuable in the context of automated nucleic acid extraction from challenging sample types like stool, where inhibitor removal and extraction efficiency directly impact assay sensitivity.
The integration of probit analysis into method validation provides a robust framework for characterizing the imprecision curve surrounding an assay's cutoff value. This is especially relevant for nucleic acid amplification tests (NAATs) used in gastroenteritis diagnostics, where traditional approaches to LoD determination that rely on blank sample analysis may not be applicable [77]. As molecular diagnostics for gastrointestinal pathogens continue to evolve, with an increasing emphasis on automated high-throughput platforms, rigorous LoD validation through probit analysis ensures reliable performance characteristics for clinical decision-making.
Theoretical Foundation of Probit Analysis
Probit analysis has its origins in toxicology and biological assays dating back to the 1940s, where it was used to characterize dose-response relationships [77]. The method operates on the principle that the cumulative probability of detection follows a sigmoidal distribution when plotted against analyte concentration. This S-shaped curve corresponds to the cumulative distribution function of a normal distribution, representing the probability of obtaining a positive result at different concentrations of the target analyte [77].
The term "probit" refers to "probability unit" and is calculated as the number of standard deviations from the mean of a normal distribution, plus a value of 5 for mathematical convenience [77]. This transformation converts the proportional data (percent positive) into a linear scale that can be analyzed using standard regression techniques. Key points on this curve include:
- C50: The concentration at which 50% of replicates test positive (probit = 5.0)
- C95: The concentration at which 95% of replicates test positive (probit = 6.64)
- C5: The concentration at which 5% of replicates test positive (probit = 3.36) [77]
For NAATs targeting gastrointestinal pathogens in stool samples, the C95 point is typically defined as the LoD, as it represents the concentration that can be detected with 95% probability [77]. This statistical framework accommodates the binary nature of qualitative test results while providing a rigorous method for estimating the detection limit across the imprecision interval.
Materials and Equipment
Research Reagent Solutions
The following table outlines essential materials and their functions for conducting probit analysis in automated nucleic acid extraction from stool samples:
Table 1: Essential Research Reagents and Materials
| Item | Function/Application |
|---|---|
| Negative stool matrix | Provides a clinically relevant, pathogen-free background for spiking experiments [57] |
| Quantified pathogen stock | Serves as the reference material for creating serial dilutions (e.g., viral, bacterial, or parasitic targets) [77] |
| Nucleic acid extraction kits | Automated extraction reagents (e.g., QIAsymphony Virus/Bacteria Midi Test Kit) for consistent nucleic acid purification [79] |
| Internal control materials | Monitors extraction efficiency and detects PCR inhibition in complex stool matrices [80] |
| PCR master mix reagents | Provides enzymes, buffers, and nucleotides for amplification; must be compatible with the extracted nucleic acid [7] |
| Positive & negative controls | Verifies assay performance and establishes baseline detection thresholds [77] |
Instrumentation Platforms
Automated nucleic acid extraction systems have demonstrated variable performance characteristics depending on the target pathogen and stool matrix composition. Comparative studies have evaluated several platforms for processing stool samples:
Table 2: Automated Nucleic Acid Extraction Systems for Stool Samples
| Instrument | Performance Characteristics | Applicable Stool Pathogens |
|---|---|---|
| MagNA Pure Compact (Roche) | Most consistent results for rotavirus RNA detection; superior inhibitor removal [7] | Rotavirus [7], Norovirus [80] |
| KingFisher Flex (ThermoFisher) | Good detection sensitivity; compatible with bead-beating for difficult-to-lyse organisms [54] | Bacterial targets, helminths [57] [54] |
| NucliSENS easyMAG (bioMérieux) | Superior precision and extraction yields for most agents; uses GuSCN for effective nuclease inactivation [81] | Bacterial biothreat agents, viruses [81] |
| QIAsymphony SP (Qiagen) | Good detection rates for norovirus RNA; integrated automated solution [80] [79] | CMV [79], Norovirus [80] |
| BioRobot M48 (Qiagen) | Reference method in comparison studies; reliable performance [79] | CMV [79] |
The selection of an appropriate extraction platform must consider the target pathogen characteristics (e.g., enveloped viruses vs. hardy helminth eggs), throughput requirements, and compatibility with downstream amplification systems [81] [7]. For robust LoD determination, the chosen platform should demonstrate consistent performance across the expected concentration range, with minimal inter-sample variability.
Experimental Protocol for LoD Determination Using Probit Analysis
Sample Preparation and Experimental Design
The following workflow outlines the complete experimental process for establishing LoD using probit analysis:
Figure 1: Experimental workflow for LoD determination using probit analysis
Step 1: Preparation of Stool Matrix
- Collect pathogen-free stool samples confirmed through comprehensive testing
- Process stool matrix to maintain consistency (homogenization, aliquoting)
- Store prepared matrix at -80°C until use [57]
Step 2: Serial Dilution Preparation
- Obtain quantified reference material (culture, synthetic, or clinical isolate)
- Prepare a minimum of 5-8 serial dilutions in negative stool matrix
- Target concentrations should bracket the expected LoD, with higher density around C50 and C95
- Include negative controls (matrix only) to confirm specificity [77] [82]
Step 3: Automated Nucleic Acid Extraction
- Process replicates (typically n=20-24 per concentration) through automated extraction
- Incorporate internal controls to monitor extraction efficiency and inhibition
- Use consistent input and elution volumes across all samples
- For difficult-to-lyse organisms (e.g., helminth eggs), include mechanical disruption such as bead beating [57] [54]
Step 4: Amplification and Detection
- Perform PCR or RT-PCR using standardized conditions
- Include appropriate positive and negative amplification controls
- Record binary results (positive/negative) for each replicate
- Document quantification cycle (Cq) values for potential secondary analysis [7] [79]
Probit Regression Analysis
Step 5: Data Analysis and LoD Calculation
- Calculate proportion positive (P) for each concentration: P = (number positive)/(total replicates)
- Convert proportions to probits using the transformation: probit = 5 + NORMSINV(P)
- Perform linear regression of probits against log10(concentration)
- Calculate C95 from the regression equation by solving for concentration when probit = 6.64 [77]
The regression equation takes the form: Probit = a + b × log10(concentration) where 'a' is the y-intercept and 'b' is the slope.
To calculate C95: log10(C95) = (6.64 - a) / b The LoD is then derived as 10^log10(C95) [77].
Practical Implementation Example
Case Study: SARS-CoV-2 LoD Determination
A multicenter study evaluating the Cepheid Xpert Xpress SARS-CoV-2 test provides a robust example of probit implementation. Researchers spiked SARS-CoV-2 virus into negative nasal pharyngeal swab matrix at seven concentrations ranging from 0.0001 to 0.0200 PFU/mL. They tested a minimum of 22 replicates at each concentration and applied probit regression analysis, which estimated the LoD at 0.005 PFU/mL. This was subsequently verified by testing 22 replicates at the next highest concentration (0.01 PFU/mL), which yielded 100% positivity [77].
Data Analysis and Interpretation
The following table illustrates a reconstructed data set from an EP17-A2 example, demonstrating the probit calculation process:
Table 3: Probit Regression Analysis for LoD Determination
| Concentration | Total Replicates | Number Positive | Proportion Positive | Probit Value |
|---|---|---|---|---|
| 10 | 100 | 8 | 0.08 | 3.58 |
| 15 | 100 | 15 | 0.15 | 3.96 |
| 20 | 100 | 29 | 0.29 | 4.45 |
| 25 | 100 | 45 | 0.45 | 4.87 |
| 30 | 100 | 61 | 0.61 | 5.28 |
| 35 | 100 | 76 | 0.76 | 5.71 |
| 40 | 100 | 87 | 0.87 | 6.13 |
| 45 | 100 | 93 | 0.93 | 6.48 |
| 50 | 100 | 97 | 0.97 | 6.88 |
For this dataset, regression analysis yields a slope of 2.062 and y-intercept of 3.353. The C95 (LoD) is calculated as follows: log10(C95) = (6.64 - 3.353) / 2.062 = 1.594 C95 = 10^1.594 = 39.2 [77]
This example highlights the importance of adequate data points across the dynamic range, particularly between C10 and C90, for reliable regression modeling.
Critical Factors in Experimental Design
Impact of Extraction Method on LoD
The choice of nucleic acid extraction methodology significantly impacts LoD determination for stool samples. Studies comparing standard versus harsh extraction protocols have demonstrated that the optimal approach varies by target organism:
Table 4: Impact of Extraction Methods on Detection Efficiency
| Extraction Method | Target Category | Effect on Detection | Key Findings |
|---|---|---|---|
| Standard silica-column based (QIAamp DNA Stool Mini Kit) | Most bacteria, viruses, protozoa | Reliable for most targets; may lack sensitivity for robust structures [57] [7] | Effective for rotavirus detection; good inhibitor removal [7] |
| Bead-beating enhanced | Helminths, Gram-positive bacteria | Significantly improved detection for 20% of helminth assays [57] [54] | Higher positive rates and lower Cq values for tough-walled organisms [57] |
| Guanidinium thiocyanate-based (easyMAG) | Broad spectrum | Superior precision and extraction yields for most agents [81] | Strong chaotropic salt enhances nuclease inactivation and cell lysis [81] |
| Microfluidic cartridge-based | Bacteria, viruses in stool | Similar or higher performance compared to conventional methods [83] | Integrated pretreatment, lysis, washing, and elution in automated format [83] |
Sensitivity of Probit Analysis to Experimental Parameters
Recent sensitivity analyses have revealed how experimental design choices affect the reliability of LoD estimates:
- Number of Concentrations: Restricting data sets to fewer concentrations lowers the estimated LLoD and widens confidence intervals [82]
- Distribution of Concentrations: Top-weighted distributions (clustered near the high end) produce less reliable estimates than centered distributions [82]
- Sample Size: Fewer than 20 replicates per concentration may compromise the precision of proportion estimates [77] [82]
- Model Fit: The Akaike information criterion (AIC) or Pearson chi-square test should assess goodness-of-fit; poor fit indicates unreliable LoD estimates [82] [79]
These findings reinforce CLSI recommendations for including a sufficient number of properly distributed concentrations in LoD studies [82].
Verification and Quality Assurance
After establishing the preliminary LoD through probit analysis, verification experiments are essential. This involves testing multiple replicates (typically 20-24) at the estimated LoD concentration and confirming ≥95% detection rate [77] [79]. Additional quality measures include:
- Testing negatives and low positives to ensure specificity
- Monitoring internal control performance across all samples
- Assessing precision through inter-run and intra-run variability [79]
- Maintaining consistent sample input and elution volumes, as these directly impact the absolute LoD [79]
For automated extraction platforms, cross-contamination studies should be performed using checkerboard patterns of high-positive and negative samples to exclude carryover [79].
Probit analysis provides a statistically rigorous framework for determining the Limit of Detection in molecular assays targeting gastrointestinal pathogens in stool samples. When properly implemented with appropriate experimental design—including sufficient replication, well-distributed concentrations, and optimized nucleic acid extraction—this method yields reliable and reproducible LoD estimates that meet regulatory requirements. The integration of automated extraction platforms enhances standardization while ensuring efficient nucleic acid purification from complex stool matrices. As molecular diagnostics continue to evolve, probit analysis remains an essential tool for establishing the analytical sensitivity required for clinical utility.
The diagnostic detection of enteric pathogens via nucleic acid amplification tests (NAATs) represents a significant advance over traditional culture methods, offering improved sensitivity and faster turnaround times [84]. However, this high sensitivity introduces a critical challenge for specificity testing: the ability to distinguish true pathogens from commensal flora and to accurately identify the causative agent in polymicrobial infections. The human gut harbors a complex ecosystem of commensal microorganisms, and their nucleic acids are co-extracted alongside pathogen targets, creating substantial risk for false-positive results or misinterpretation of findings [84] [85]. This application note addresses this challenge within the context of automated nucleic acid extraction from stool samples, providing a systematic framework for evaluating and ensuring assay specificity.
The core of this challenge lies in the fundamental difference between NAATs and traditional culture. While NAATs detect the presence of target genetic material, they do not discriminate between viable pathogens, non-viable organisms, or commensal strains that may be present without causing disease [84]. This is particularly problematic for organisms like enteroaggregative E. coli (EAEC) and enteropathogenic E. coli (EPEC), which are frequently identified by multiplex panels but whose clinical significance in symptomatic patients can be ambiguous [84]. Furthermore, the presence of commensal fungi, such as Candida albicans, has been shown to influence bacterial virulence, adding another layer of complexity to diagnostic interpretation [86]. Therefore, rigorous specificity testing that evaluates cross-reactivity with commensal flora and co-infecting pathogens is not merely a regulatory formality but an essential component of robust assay design and implementation.
Experimental Design for Comprehensive Specificity Testing
A robust specificity testing protocol must assess an assay's performance against two key backgrounds: a representative panel of commensal organisms and complex clinical matrices containing multiple potential pathogens.
Commensal Flora Panel Construction
The first step involves creating a well-characterized panel of commensal organisms that are typically present in stool samples. The composition of this panel should be informed by molecular studies of microbial populations, which have revealed a diverse bacterial landscape on even non-gut surfaces, dominated by coagulase-negative staphylococci but also containing atypical species like Rhodococcus erythropolis and Klebsiella oxytoca [87]. The panel should include:
- Commensal Bacteria: A diverse selection of organisms from phyla such as Firmicutes, Bacteroidetes, and Actinobacteria, which constitute the predominant gut flora [85].
- Commensal Fungi: Species such as Candida albicans, a common gut commensal whose presence has been demonstrated to modulate the virulence of bacterial pathogens like Salmonella Typhimurium [86].
- Near-Neighbors: Non-pathogenic species that are phylogenetically closely related to the target pathogens, posing a higher risk for cross-reactivity.
This panel is used to challenge the NAAT following automated nucleic acid extraction to verify that no non-target organisms generate a false-positive signal for any of the pathogens on the panel.
Cross-Reactivity Testing with Polymicrobial Samples
The second, more complex, step involves testing the assay's ability to correctly identify a target pathogen in the presence of other potential pathogens. This is crucial because co-infections do occur, and the diagnostic platform must be able to identify the true causative agent(s). The experimental design should:
- Spike Clinical Matrices: Introduce a known quantity of a target pathogen (e.g., Salmonella) into a standardized stool matrix that either naturally contains or has been spiked with non-target organisms (e.g., Campylobacter, Shigella, norovirus).
- Vary Relative Abundances: Test across a range of pathogen-to-commensal ratios to determine the assay's limit of accurate detection for the target amidst background interference.
- Evaluate Detection Specificity: Confirm that the detection of the target pathogen remains specific and that its presence does not inadvertently enhance or suppress the detection of other targets.
Table 1: Key Experimental Parameters for Specificity Testing
| Parameter | Description | Application in This Context |
|---|---|---|
| Commensal Panel | A curated collection of non-pathogenic organisms typically found in the gut microbiome. | Challenge the assay post-automated extraction to confirm no cross-detection. |
| Polymicrobial Samples | Samples containing mixtures of multiple pathogens at varying concentrations. | Validate the assay's ability to correctly identify all relevant targets in a co-infection scenario. |
| Sample Input Volume | The volume of stool sample processed by the automated extractor. | Optimize for sufficient pathogen yield while minimizing PCR inhibitors. |
| Automated Extraction Method | The specific platform and chemistry used for nucleic acid co-extraction. | Ensure efficient lysis of diverse pathogens (bacterial, viral, parasitic) and inhibitor removal. |
Detailed Protocols
Protocol 1: Specificity Testing Against a Commensal Flora Panel
This protocol evaluates the potential for cross-reactivity between pathogen-specific detection assays and nucleic acids from commensal organisms.
Materials & Reagents:
- Automated Nucleic Acid Extraction System: e.g., BioRobot M48, MagNA Pure, or LunaDx Pro system [88] [89].
- Extraction Kits: Compatible with the automated system for total nucleic acid or pathogen-specific extraction from stool.
- Commensal Strain Panel: Cultured isolates or quantified nucleic acids from ~20-30 representative commensal bacteria and fungi.
- Negative Stool Matrix: Stool from healthy volunteers, confirmed negative for the target pathogens by a validated method.
- NAAT Reagents: Master mix, primers, and probes for all target pathogens on the diagnostic panel.
Procedure:
- Sample Preparation: Suspend each commensal organism to a high concentration (e.g., ≥ 10^6 CFU/mL for bacteria, ≥ 10^5 spores/mL for fungi) in the negative stool matrix.
- Automated Nucleic Acid Extraction:
- Load the spiked stool samples onto the automated extraction platform.
- Use a sample input volume of 200-300 µL, as optimized for the system [89].
- Execute the extraction protocol according to the manufacturer's instructions. The protocol should include a robust lysis step (e.g., using chaotropic salts and detergents) to ensure efficient disruption of all organism types [90].
- Elute the purified nucleic acids in a volume of 50-150 µL.
- Pathogen Detection:
- Test the eluted nucleic acids using the multiplex NAAT platform (e.g., PCR, FilmArray, Luminex).
- Include appropriate positive controls (pure target pathogen nucleic acid) and negative controls (nuclease-free water and unspiked negative stool matrix).
- Analysis:
- Record any signal detected for the pathogen targets. A true negative result shows no detection signal from the commensal-only samples.
- If cross-reactivity is observed, investigate sequence homology at the primer/probe binding sites and redesign the assays if necessary.
Protocol 2: Specificity and Interference Testing in Polymicrobial Samples
This protocol assesses the assay's accuracy for identifying a target pathogen in the presence of other pathogens, simulating a complex clinical co-infection.
Materials & Reagents:
- All materials from Protocol 1.
- Target Pathogen Stock: Quantified culture or nucleic acid of the primary pathogen of interest (e.g., Salmonella Typhimurium).
- Interfering Organism Stocks: Quantified cultures or nucleic acids of other pathogens (e.g., Clostridium difficile, Giardia lamblia, norovirus).
Procedure:
- Experimental Matrix Creation:
- Prepare a dilution series of the target pathogen in negative stool matrix, spanning the assay's limit of detection (LoD) to 100x LoD.
- Spike a constant, high concentration (e.g., 10x the target's concentration) of one or more interfering organisms into each dilution of the target pathogen.
- Automated Co-extraction and Multiplex Detection:
- Process all samples through the automated nucleic acid extraction and multiplex NAAT platform as described in Protocol 1.
- Data Collection and Analysis:
- For each sample, record the cycle threshold (Ct) value or qualitative result for the target pathogen and all other pathogens in the panel.
- Compare the detection of the target pathogen in the presence vs. absence of the interfering organisms.
- A robust assay will show no statistically significant difference in the Ct value or qualitative result for the target pathogen, and will correctly identify (or not identify) the interfering organisms as appropriate.
Table 2: Example Results from a Specificity and Interference Study
| Target Pathogen | Interfering Organism | Target Ct (Alone) | Target Ct (with Interferent) | Interferent Detected? | Conclusion |
|---|---|---|---|---|---|
| Salmonella | Campylobacter jejuni | 25.1 | 25.4 | Yes (Correct) | No Interference |
| Shigella | Norovirus GI/GII | 28.5 | 28.3 | Yes (Correct) | No Interference |
| Giardia | Cryptosporidium | 30.2 | Undetected | Yes (Correct) | Significant Interference |
| Clostridium difficile | Candida albicans | 26.8 | 26.9 | No (Correct) | No Interference |
The Scientist's Toolkit: Key Research Reagent Solutions
The following reagents and materials are critical for executing the described specificity evaluations.
Table 3: Essential Research Reagents for Specificity Evaluation
| Reagent/Material | Function | Key Considerations |
|---|---|---|
| Automated Extraction Kit | Purifies nucleic acids from complex stool samples. | Must be validated for simultaneous extraction of DNA and RNA from a broad range of pathogens and commensals [88] [90]. |
| Characterized Commensal Strain Panel | Serves as the negative control panel for cross-reactivity testing. | Should be phylogenetically diverse and include organisms with high sequence similarity to targets. |
| Quantified Pathogen Standards | Provides known concentrations of target and interfering pathogens for spiking studies. | Essential for determining LoD and for creating accurate polymicrobial models; available from ATCC and other biological resource centers. |
| Stool Collection & Transport Kit | Preserves sample integrity from collection to processing. | Kits with DNA/RNA stabilizing solutions (e.g., DESS, Zymo DNA Shield) prevent microbial population shifts and nucleic acid degradation at room temperature [85]. |
| Multiplex NAAT Master Mix | Amplifies and detects multiple pathogen-specific targets in a single reaction. | Must be resistant to carry-over inhibitors from stool and compatible with the extracted nucleic acid eluent. |
Workflow and Data Interpretation
The following diagram illustrates the logical workflow for conducting and interpreting specificity tests, guiding the researcher from experimental setup to final assay validation.
Specificity Testing Workflow
Interpreting the results requires careful clinical correlation. A positive NAAT result does not always indicate active disease, as the test may detect nucleic acid from non-viable organisms or asymptomatic carriage [84]. This is a known limitation of NAATs compared to culture, which provides a viable isolate. Therefore, the results of these specificity tests must be integrated with clinical data. For instance, the detection of a pathogen in a stool sample from a patient with a compatible clinical syndrome (e.g., Salmonella in a patient with acute inflammatory diarrhea) strongly suggests a causative role. In contrast, the detection of a pathogen like EPEC in an asymptomatic patient may be of uncertain significance [84]. Understanding these nuances is critical for researchers developing assays and for clinicians interpreting the results.
Ensuring the specificity of multiplex NAATs in the complex environment of stool samples is a critical endeavor. By implementing the systematic experimental design and detailed protocols outlined in this application note—evaluating cross-reactivity with commensal flora and performance in polymicrobial samples—researchers and developers can robustly validate their automated nucleic acid extraction and detection platforms. This rigorous approach to specificity testing is fundamental to delivering accurate, reliable diagnostic results that can effectively guide patient treatment and public health responses.
The efficiency of nucleic acid extraction is a critical determinant of success in molecular diagnostics and research, particularly when working with complex sample matrices such as stool. The choice of extraction methodology can significantly impact downstream analytical results, including sensitivity, reproducibility, and suitability for automation. This application note provides a detailed comparative analysis of three dominant nucleic acid purification technologies: traditional silica spin columns, magnetic beads, and novel nanoparticles. Framed within the context of automated extraction from stool samples—a sample type known for its high inhibitor content and heterogeneity—this document delivers structured performance data, detailed experimental protocols, and practical guidance for researchers and drug development professionals.
The following table summarizes the core characteristics and performance metrics of the three extraction methods, providing a baseline for technology selection.
Table 1: Head-to-Head Comparison of Nucleic Acid Extraction Technologies
| Feature | Silica Spin Columns | Magnetic Beads | Novel Nanoparticles |
|---|---|---|---|
| Core Principle | Silica membrane in a column binds nucleic acids under chaotropic conditions [91] | Silica- or carboxyl-coated magnetic beads bind nucleic acids for reversible immobilization [92] [93] | Engineered magnetic nanoparticles with optimized coatings for enhanced binding [94] [27] |
| Typical Binding Capacity | Varies with membrane size | High, scalable with bead volume [94] | Very high; one study reported ~96% binding for high-input DNA using 50 µL beads [94] |
| Typical Yield/Recovery | Variable; can be lower for short nucleic acids [91] | High; ~90-110% recovery rates reported for viral RNA [95] | High-yield; one method reports eluting "nearly all" nucleic acid in sample [94] |
| Processing Time | ~15-40 minutes [95] [91] [94] | Rapid; <5 minutes for manual, ~40 min for some automated kits [96] [91] [94] | Very rapid; 6-7 minutes for full protocol [94] |
| Automation Compatibility | Moderate (requires centrifuges or vacuum manifolds) [91] | Excellent (inherently suited for liquid handlers) [96] [93] [97] | Excellent (compatible with tip-based and automated systems) [94] |
| Throughput | Good with 96-well plate formats [91] | Excellent for high-throughput (simultaneous 96-well processing) [96] [92] | Promising for high-throughput applications [92] |
| Cost Consideration | Costly per sample for commercial kits [92] [91] | Cost-effective for large-scale use; low per-sample cost [92] | Highly cost-effective, especially with open-source synthesis [92] |
| Key Advantage | Simplicity, well-established protocols | Speed, automation-friendliness, no centrifugation needed [91] [93] | High binding efficiency, rapid kinetics, customizability |
Detailed Experimental Protocols
The following sections provide detailed methodologies for benchmarking these extraction technologies, with a focus on application to stool samples.
Protocol: Automated Nucleic Acid Extraction from Stool Samples Using Magnetic Beads
This protocol is adapted for a high-throughput automated system, such as the Insta NX Mag 16Plus or similar platforms [96].
Research Reagent Solutions & Materials
- Lysis Buffer (LBB): Typically containing guanidine hydrochloride (4 M), Triton X-100 (1.5%), EDTA (27.5 mM), Tris-Cl (pH 7.4), and 50% ethanol. Guanidine is a chaotropic salt that denatures proteins and facilitates nucleic acid binding to silica [95] [94].
- Wash Buffer: Usually 70-80% ethanol solution, used to remove salts, proteins, and other impurities without eluting the nucleic acids [91] [93].
- Elution Buffer (EB): Low-salt buffer such as TE buffer or nuclease-free water. Some optimized protocols use a weak base like 2 mM NaOH to disrupt silica-nucleic acid interactions [95] [94].
- Proteinase K: For digesting proteins and breaking down complex stool structures.
- Silica-Coated Magnetic Beads: Commercially available or synthesized in-house [92] [93].
- Automated Nucleic Acid Extractor: e.g., Insta NX Mag 16Plus, KingFisher, or similar magnetic particle processor [96].
- Deep-well 96-well plates compatible with the extraction system.
Workflow Steps
Protocol: High-Yield Extraction via "Tip-Based" Method (SHIFT-SP)
This manual, rapid protocol demonstrates the optimization of magnetic bead use for maximum yield and can be a reference for automation development [94].
Key Materials
- Magnetic Beads: Silica-coated magnetic beads (e.g., 30-50 µL per sample for high input) [94].
- Optimized Lysis/Binding Buffer (LBB): Guanidine-based buffer, pH adjusted to ~4.1 to enhance nucleic acid binding by reducing electrostatic repulsion [94].
- Low-salt Elution Buffer: Pre-warmed to 62°C to increase elution efficiency [94].
- Fixed-volume pipette (e.g., 1 mL) or a repetitive pipettor.
Workflow Steps
- Lysis: Homogenize 100-200 mg of stool sample in 1 mL of LBB. Incubate at 62°C for 3-5 minutes to facilitate complete lysis.
- Tip-Based Binding: Add 30-50 µL of magnetic bead suspension to the lysate. Instead of vortexing or orbital shaking, aspirate and dispense the entire mixture repeatedly for 2 minutes using a pipette set to 80% of the total volume. This "tip-based" mixing rapidly exposes beads to nucleic acids, significantly improving binding kinetics and yield [94].
- Magnetic Separation: Place the tube on a magnetic rack for 30-60 seconds until the solution clears. Carefully pipette off and discard the supernatant.
- Washing: With the tube still on the magnet, add 500 µL of Wash Buffer (70-80% ethanol). Gently tap the tube to resuspend the beads. Let it stand for 30 seconds, then pipette off the wash supernatant. Repeat this wash step a second time.
- Drying: Air-dry the bead pellet for about 2 minutes at room temperature to ensure complete ethanol evaporation.
- Elution: Remove the tube from the magnetic rack. Add 50-100 µL of pre-warmed Elution Buffer and pipette mix thoroughly. Incubate at 62°C for 1-2 minutes.
- Final Separation: Place the tube back on the magnetic rack. After 1 minute, transfer the supernatant containing the purified nucleic acids to a clean tube.
Performance Data and Analysis
Quantitative data from comparative studies provides critical insights for decision-making. The following table consolidates key performance indicators from recent literature.
Table 2: Quantitative Performance Metrics from Comparative Studies
| Extraction Method | Sample Type | Processing Time | Nucleic Acid Recovery / Yield | Purity (A260/A280) | Key Finding |
|---|---|---|---|---|---|
| Silica Pipette Tip Column [95] | Nasopharyngeal swab, Saliva | < 3 minutes | 90-110% (viral RNA) | Implied high (successful in LAMP) | Equipment-free, comparable to traditional silica columns but faster. |
| Magnetic Beads (Automated) [96] | Plasma, Serum, Swabs | ~40 minutes (for 16 samples) | Detection of HBV down to 2.39 IU/μL | High (validated for qPCR) | High concordance (>99%) with commercial kits for HIV, HBV, HCV. |
| Magnetic Beads (High-Yield) [94] | Bacterial culture, Spiked whole blood | 6-7 minutes | ~96% of input DNA bound | High (suitable for WGA & sequencing) | "Tip-based" binding and low pH LBB dramatically increase yield and speed. |
| Commercial Column Kit [94] | Bacterial culture | 25 minutes | ~50% of SHIFT-SP DNA yield | Not specified | Benchmark method; slower and lower yielding than optimized bead protocols. |
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents and Materials for Automated Nucleic Acid Extraction
| Item | Function / Description | Example / Note |
|---|---|---|
| Chaotropic Salt | Disrupts hydrogen bonding, denatures proteins, and enables nucleic acid binding to silica surfaces. | Guanidine hydrochloride or guanidine thiocyanate [95] [91] [94]. |
| Silica-Coated Magnetic Beads | Solid phase for reversible nucleic acid binding and immobilization via magnetic fields. | Commercially available Sera-Mag beads or laboratory-synthesized beads [92] [93]. |
| Proteinase K | Digests proteins and nucleases that could degrade the target nucleic acid. | Critical for complex samples like stool to break down cellular and fecal material. |
| Ethanol Wash Buffer | Removes salts, solvents, and other contaminants from the bead-nucleic acid complex. | Typically 70-80% concentration [91] [93]. |
| Low-Salt Elution Buffer | Disrupts the interaction between the nucleic acid and the silica surface, releasing pure NA into solution. | TE buffer or nuclease-free water; pre-warming to 62°C can increase yield [94]. |
| Automated Extraction System | Instrument for high-throughput, hands-free nucleic acid purification using magnetic beads. | Insta NX Mag 16Plus, ThermoFisher KingFisher, Hamilton STAR [96] [91]. |
The comparative data and protocols presented herein clearly indicate a paradigm shift towards magnetic bead-based technologies for automated, high-throughput nucleic acid extraction, especially from challenging samples like stool. The superior speed, ease of automation, and high recovery rates of magnetic beads offer tangible advantages over traditional silica columns [95] [96] [94].
For stool sample research specifically, where inhibitor removal and yield are paramount, the optimized "tip-based" magnetic bead protocol (SHIFT-SP) is particularly compelling. Its use of low-pH binding buffer and active pipette mixing addresses key inefficiencies, resulting in a rapid, high-yield workflow suitable for downstream sensitive applications like whole genome amplification and sequencing [94].
While silica columns remain a reliable and simple option for low-throughput applications, and novel nanoparticles show promise for future customizability and performance gains, magnetic beads currently represent the optimal solution for automated, high-performance nucleic acid extraction in drug development and clinical research. Future work in this field will likely focus on further integrating these optimized bead-based extraction protocols with amplification and detection steps in fully closed, automated systems to minimize contamination and maximize throughput in diagnostic pipelines [98].
The reliability of molecular diagnostics is fundamentally rooted in the quality of the pre-analytical phase, with nucleic acid extraction efficiency serving as a critical determinant of diagnostic sensitivity [99]. This correlation is particularly crucial when working with complex and inhibitor-rich samples such as stool, which present significant challenges for robust molecular testing [12] [6]. Efficient extraction methodologies must not only maximize nucleic acid yield but also effectively eliminate PCR inhibitors commonly found in clinical specimens, thereby ensuring the accuracy and reliability of downstream diagnostic applications [100].
Automated extraction platforms have emerged as essential tools for standardizing this process, reducing human error, and improving reproducibility across patient cohorts [12] [101]. The integration of automated systems in clinical laboratories has demonstrated enhanced performance in high-throughput settings, providing consistent results that are vital for both routine diagnostics and large-scale research studies [101] [102]. This application note explores the direct relationship between extraction efficiency and diagnostic sensitivity, focusing specifically on automated nucleic acid extraction from stool samples within the broader context of patient cohort studies.
Performance Metrics: Quantitative Comparison of Extraction Systems
The evaluation of nucleic acid extraction systems requires comprehensive assessment across multiple performance parameters. The following tables summarize key quantitative metrics derived from comparative studies of automated extraction platforms, highlighting their impact on diagnostic sensitivity in clinical settings.
Table 1: Performance Comparison of Automated Nucleic Acid Extraction Systems for Complex Samples
| Extraction System | Sample Type | Extraction Efficiency | Inhibitor Removal | Concordance with Reference Methods | Precision (CV) |
|---|---|---|---|---|---|
| EZ2 PowerFecal Pro [12] | Stool | High DNA yields | Patent-pending Inhibitor Removal Technology | High | <5% |
| PANA HM9000 [101] | Plasma, swabs | 100% detection rate for low concentrations | Effective inhibitor removal | 100% positive, negative, and overall concordance | <5% intra- and inter-assay |
| NucliSens easyMAG [102] | Clinical specimens | R²=0.99 for linearity | Reduced inhibition | 95.7% for CMV, 100% for EBV | Low inter- and intrarun variability |
| Five-Minute Extraction (FME) [103] | Respiratory samples | Superior RNA concentration and purity | Effective removal of PCR inhibitors | 95.43% total coincidence rate with standard methods | High reproducibility |
Table 2: Impact of Pre-analytical Conditions on Cell-free DNA Yield from Blood Collection Tubes
| Blood Collection Tube | cfDNA Yield at 0h (ng/mL) | cfDNA Yield at 48h (ng/mL) | cfDNA Yield at 168h (ng/mL) | Stability Profile |
|---|---|---|---|---|
| K₂EDTA [104] | 2.41 | 7.39 | 68.19 | Significant increase over time |
| Streck [104] | 2.74 | 2.63 | 2.38 | Minimal decrease (13.1%) |
| PAXgene [104] | 1.66 | 1.72 | 2.48 | Moderate increase (49.4%) |
| Norgen [104] | 0.76 | 0.75 | 0.76 | Highly stable |
The data from these comparative analyses demonstrate that automated extraction systems consistently provide high efficiency and reproducibility across diverse sample types. The EZ2 PowerFecal Pro system specifically addresses challenges associated with stool samples through specialized inhibitor removal technology, while platforms like the PANA HM9000 show exceptional concordance rates in clinical validation studies [12] [101]. The impact of pre-analytical conditions further highlights the importance of standardized collection and processing protocols for maintaining sample integrity and ensuring reliable diagnostic results [104].
Experimental Protocols
Automated Nucleic Acid Extraction from Stool Samples Using the EZ2 PowerFecal Pro System
Principle: This protocol enables the automated isolation of microbial DNA, RNA, or total nucleic acids from stool samples using the EZ2 Connect platform with integrated Inhibitor Removal Technology [12].
Materials:
- EZ2 PowerFecal Pro DNA/RNA Kit (QIAGEN, cat. no. 954634)
- Fresh or appropriately preserved stool samples
- EZ2 Connect Instrument
- PowerProtect DNA/RNA Reagent (for sample preservation)
- RNase A (17,500 U) (cat. no. 19101) for DNA-only extraction
- RNase-free DNase Set (cat. no. 79254) for RNA-only extraction
- Tissuelyser III with appropriate adapters (for sample homogenization)
Procedure:
- Sample Collection and Preservation:
- Collect 50-100 mg of stool sample using appropriate collection devices.
- For stabilization, use PowerProtect DNA/RNA reagent if immediate processing is not possible.
- Process samples as quickly as possible after collection to optimize nucleic acid quality.
Sample Lysis and Homogenization:
- Transfer 50-100 mg of stool to a tube containing lysis buffer.
- Add phenol chloroform isoamyl alcohol for inhibitor depletion and RNase deactivation.
- Homogenize using Tissuelyser III with appropriate adapters for complete disruption.
Automated Extraction on EZ2 Connect:
- Transfer the eluate from the lysis step to the EZ2 Connect instrument.
- Select the appropriate protocol based on desired output (DNA-only, RNA-only, or total nucleic acids).
- For DNA-only extraction, the instrument automatically performs RNase A digestion.
- For RNA-only extraction, the instrument automatically performs DNase digestion.
- The system utilizes magnetic particle technology for nucleic acid binding, washing, and elution.
- Process up to 24 samples in a single run.
Elution and Storage:
- Elute nucleic acids in the provided elution buffer.
- Assess concentration and purity using spectrophotometric methods.
- Store extracted nucleic acids at -20°C to -80°C for long-term preservation.
Downstream Applications: The extracted nucleic acids are suitable for RT-PCR, qPCR, dPCR, and next-generation sequencing applications, including RNA-seq and metatranscriptome analysis [12].
Comprehensive Performance Validation of Automated Extraction Systems
Principle: This protocol outlines a standardized framework for validating the performance of automated nucleic acid extraction systems in clinical settings, based on CLSI guidelines [101].
Materials:
- Automated molecular detection system (e.g., PANA HM9000)
- Clinical samples with known pathogen status
- WHO international standards and national reference materials
- Appropriate detection kits (e.g., EBV DNA, HCMV DNA, RSV RNA detection kits)
- Quantitative PCR instrumentation
Procedure:
- Concordance Rate Assessment:
- Collect residual clinical samples (e.g., plasma, oropharyngeal swabs).
- Test samples using both the automated system and reference method.
- Calculate positive, negative, and overall concordance rates according to CLSI EP12 guidelines.
Accuracy Evaluation:
- Prepare dilution series from WHO international standards.
- Test each concentration in triplicate with multiple extractions.
- Compare mean detection values with theoretical clinical values per CLSI EP09.
Linearity Assessment:
- Create five concentration gradients from reference standards.
- Analyze linear correlation coefficient (|r|) according to CLSI EP06.
- Establish the measuring interval where results are proportional to analyte concentrations.
Precision Determination:
- Perform intra-assay and inter-assay precision testing.
- Calculate coefficients of variation (CV) with target of <5%.
- Follow CLSI EP05 guidelines for experimental design.
Limit of Detection (LoD) Establishment:
- Test serial dilutions of standardized materials.
- Determine the lowest concentration consistently detected.
- Follow CLSI EP17 guidelines for LoD verification.
Interference and Cross-reactivity Testing:
- Assess potential interference from common substances.
- Evaluate cross-reactivity with related pathogens.
- Follow CLSI EP07 standards for validation.
Carryover Contamination Assessment:
- Process high-positive samples followed by negative controls.
- Monitor for any signal in negative samples indicating carryover.
- Document any contamination events and frequencies.
Validation Criteria: The system should demonstrate high concordance rates (>95%), excellent precision (CV<5%), appropriate linearity (|r|≥0.98), and minimal interference or cross-reactivity to be considered suitable for clinical implementation [101].
Workflow Visualization: Automated Nucleic Acid Extraction and Diagnostic Correlation
The following diagram illustrates the complete workflow from sample collection through diagnostic interpretation, highlighting critical control points where extraction efficiency impacts ultimate diagnostic sensitivity:
Diagram Title: Automated Nucleic Acid Extraction to Diagnostic Correlation Workflow
This workflow emphasizes the sequential process where optimal performance at each stage is essential for maintaining diagnostic accuracy. The critical control points, particularly inhibitor removal and quality assessment, serve as key determinants of the final clinical correlation metrics including sensitivity, specificity, and predictive values.
The Scientist's Toolkit: Essential Research Reagent Solutions
The following table details key reagents and materials essential for successful implementation of automated nucleic acid extraction from stool samples, along with their specific functions in the experimental workflow:
Table 3: Essential Research Reagent Solutions for Automated Nucleic Acid Extraction
| Reagent/Material | Function | Application Notes |
|---|---|---|
| EZ2 PowerFecal Pro DNA/RNA Kit [12] | Automated isolation of microbial DNA, RNA, or total nucleic acids | Pre-filled cartridges minimize human errors; compatible with EZ2 Connect platform |
| Inhibitor Removal Technology [12] | Specialized chemistry for removing PCR inhibitors from complex samples | Critical for stool samples with high inhibitor content; improves downstream applications |
| PowerProtect DNA/RNA Reagent [12] | Stabilizes stool samples at room temperature | Essential when immediate processing is not possible; preserves nucleic acid integrity |
| Magnetic Beads (Silica-coated) [21] | Nucleic acid binding through silica surface in chaotropic salt conditions | Enable efficient capture and release; gentle separation minimizes shearing |
| Lysis Buffers (Guanidine Thiocyanate) [103] | Cell disruption and nucleic acid release | Often combined with sodium citrate, sarkosyl, and DTT for enhanced efficiency |
| Wash Buffers (Glycerin/Ethanol) [103] | Removal of contaminants and inhibitors | Optimized 50:50 glycerin/ethanol solution provides effective washing in abbreviated cycles |
| Elution Buffers (Tris-HCl/EDTA) [103] | Final recovery of purified nucleic acids | Low-salt solutions facilitate elution from silica surfaces; maintain nucleic acid stability |
| Enzymatic Digestion Reagents (RNase A, DNase) [12] | Selective removal of unwanted nucleic acids | Required for DNA-only or RNA-only extraction; purchased separately |
These specialized reagents form the foundation of robust automated extraction systems, each playing a specific role in overcoming the unique challenges presented by complex sample matrices like stool. Proper selection and application of these solutions directly impact the efficiency of nucleic acid recovery and the elimination of substances that could interfere with downstream diagnostic applications.
The direct correlation between nucleic acid extraction efficiency and diagnostic sensitivity underscores the critical importance of optimized, automated extraction methodologies in clinical molecular diagnostics. For challenging sample types such as stool, specialized systems incorporating robust inhibitor removal technologies and standardized protocols ensure consistent performance across diverse patient cohorts. The quantitative data and methodological frameworks presented in this application note provide researchers and clinicians with evidence-based guidance for implementing automated extraction systems that maximize diagnostic sensitivity and reliability in both routine practice and research settings.
Within the broader thesis investigating automated nucleic acid extraction from stool samples, the assessment of methodological precision is paramount. For research outcomes to be credible and translatable to drug development pipelines, scientists must ensure that results are not only accurate but consistently reproducible across different runs, days, and operators [105] [101]. This application note provides a detailed protocol for a rigorous reproducibility assessment, framed within the specific context of automated nucleic acid extraction from complex stool matrices. The precision of an assay, defined as the closeness of agreement between independent measurement results obtained under stipulated conditions, is solely related to random error and is distinct from trueness or accuracy [105]. In high-throughput research and clinical settings, automated systems are marketed to reduce the inter-sample variation inherent in manual processing [5]. Verifying these claims through a structured assessment of inter-assay (intermediate precision) and intra-assay (repeatability) variability is a critical step in method validation, ensuring that subsequent microbiome, pathogen detection, or genomic data are robust and reliable [5] [101].
Core Concepts: Inter-assay vs. Intra-assay Precision
Precision evaluation must dissect the different components of variability. The terminology below is aligned with international standards, including Clinical and Laboratory Standards Institute (CLSI) guidelines [105] [106].
- Intra-Assay Precision (Repeatability): Expresses the precision under conditions where the same sample is analyzed in multiple replicates (e.g., duplicates) using the same measurement procedure, same operators, same measuring system, same operating conditions, and same location over a short period of time, typically within a single run or day [106] [107]. This measures the smallest possible variation an assay can achieve and is often quantified by the intra-assay coefficient of variation (CV%) [107].
- Inter-Assay Precision (Intermediate Precision): Expresses the within-laboratory precision obtained over a longer period (e.g., several days or weeks) and incorporates more sources of variation than repeatability. These can include different analysts, different calibrants, different batches of reagents, and different instruments [105] [106]. Its value, expressed as standard deviation or CV%, is typically larger than the repeatability standard deviation because it accounts for more random effects [106].
- Reproducibility: Represents the highest level of variability, referring to precision between measurement results obtained at different laboratories [105] [106]. While critical for method standardization, it is not always required for single-lab validation.
The relationship between these concepts and the factors they encompass is visualized in the following workflow.
Experimental Protocol for Precision Assessment
This protocol is adapted from CLSI guidelines EP05-A2 and EP15-A2, tailored for validating automated nucleic acid extraction from stool samples [105].
Experimental Design and Sample Preparation
- Materials: Use at least two levels of pooled human stool samples (e.g., from healthy volunteers) or a mock microbial community standard (e.g., ZymoBIOMICS Microbial Community Standard) to assess precision across the analytical range [105] [5]. These materials should be different from the daily quality control samples.
- Stabilization: Preserve stool samples immediately upon collection using a commercial preservation reagent (e.g., DNA/RNA Shield) and store at -80°C until extraction to stabilize nucleic acids [5] [12].
- Study Design (EP15-A2 for Manufacturer Claim Verification):
- Duration: 5 days.
- Replicates: 3 replicates per sample level per day.
- Total Analyses: 5 days × 3 replicates = 15 data points per sample level [105].
- Study Design (EP05-A2 for Comprehensive In-House Validation):
- Duration: 20 days.
- Runs: 2 runs per day, separated by at least 2 hours.
- Replicates: Duplicate analyses in each run.
- Total Analyses: 20 days × 2 runs × 2 replicates = 80 data points per sample level [105].
- Incorporating Operators: To assess the impact of different operators as part of intermediate precision, involve at least two trained analysts who independently perform the extraction and analysis on different days [105] [106].
- Downstream Analysis: Quantify extracted nucleic acids using a fluorometer (e.g., Qubit) and assess quality via spectrophotometry (e.g., NanoDrop). Confirm precision with a downstream application such as qPCR for a specific target or 16S rRNA gene amplicon sequencing [5].
Data Analysis and Calculation of Precision
The following calculations should be performed for each level of tested material.
Step 1: Calculate the Mean and Standard Deviation.
- Intra-Assay (Repeatability) Standard Deviation (sr): Calculate the standard deviation of the replicates within each run/day, then pool the results [105]. A common approach for duplicate data is to calculate the CV for each duplicate pair and then report the average CV [107].
- Inter-Assay (Intermediate Precision) Standard Deviation (sl): Also known as within-laboratory precision, this encompasses both within-run and between-run/day variability. It can be derived using analysis of variance (ANOVA) components [105].
Step 2: Calculate the Coefficient of Variation (CV%). The CV is a dimensionless number that allows for comparison across different concentrations.
CV% = (Standard Deviation / Mean) × 100[105] [107].Step 3: Evaluate Results. Compare the calculated repeatability (sr) and within-laboratory precision (sl) to the manufacturer's claims or pre-defined acceptance criteria (e.g., based on user requirements). If the observed values are less than or equal to the claims, the performance is verified. If they are larger, a statistical test (e.g., Chi-square test) is needed to determine if the difference is significant [105].
Case Study & Data Presentation
A recent study comparing automated nucleic acid extractors for fecal microbiota research provides a relevant example of precision assessment in practice [5].
Performance Comparison of Automated Extractors
Table 1: DNA Yield and Quality from Automated Nucleic Acid Extractors on Stool Samples (adapted from [5])
| Extraction System | Bead-Beating | Average DNA Yield (ng/µL)* | Purity (A260/A280)* | Inter-Sample Variability (CV%) |
|---|---|---|---|---|
| Genepure Pro | Yes | High | 1.85 - 1.95 | <5% |
| Maxwell RSC 16 | Yes | High | 1.80 - 1.92 | <5% |
| KingFisher Apex | Yes | Moderate | 1.82 - 1.90 | <5% |
| Manual (Column-based) | Yes | Moderate | 1.78 - 1.88 | 5-10% |
*Representative ranges. Actual values are sample-dependent.
The study demonstrated that all three automated extractors showed low inter-sample variability (CV <5%), outperforming manual extraction which exhibited higher variability (CV 5-10%) [5]. This highlights the improved reproducibility offered by automation.
Exemplary Precision Data from a Clinical System
A comprehensive evaluation of a high-throughput automated molecular detection system (PANA HM9000) demonstrated exceptional precision for pathogen detection, as summarized below [101].
Table 2: Inter- and Intra-Assay Precision of an Automated Molecular Detection System (adapted from [101])
| Analyte | Type of Test | Intra-Assay Precision (CV%) | Inter-Assay Precision (CV%) | Acceptance Criterion (CV%) |
|---|---|---|---|---|
| EBV DNA | Quantitative | < 5% | < 5% | < 5% |
| HCMV DNA | Quantitative | < 5% | < 5% | < 5% |
| RSV RNA | Qualitative | - | - | - |
| Sample Type | Concentration Level | Intra-Assay CV% | Inter-Assay CV% |
|---|---|---|---|
| Plasma (EBV DNA) | Low (e.g., 50 IU/mL) | 3.2% | 4.1% |
| Plasma (EBV DNA) | High (e.g., 50,000 IU/mL) | 2.1% | 3.5% |
| Plasma (HCMV DNA) | Low (e.g., 100 IU/mL) | 3.8% | 4.3% |
| Plasma (HCMV DNA) | High (e.g., 100,000 IU/mL) | 1.9% | 2.8% |
For qualitative tests, precision is often assessed via positive/negative concordance rates, which were 100% in this study [101].
The Scientist's Toolkit: Essential Research Reagent Solutions
The following reagents and instruments are critical for executing a robust precision assessment in automated nucleic acid extraction from stool samples.
Table 3: Essential Materials for Automated Nucleic Acid Extraction from Stool
| Item Category | Specific Examples | Function & Importance in Precision |
|---|---|---|
| Automated Extractors | KingFisher Apex, Maxwell RSC 16, Bioer GenePure Pro, EZ2 Connect [5] [12] | Magnetic bead-based platforms that automate binding, washing, and elution. Minimize human error and inter-operator variability, directly improving reproducibility [5] [108]. |
| Extraction Kits | MagMAX Microbiome Ultra Kit, Maxwell RSC Fecal Microbiome DNA Kit, MagaBio Fecal Pathogens DNA Purification Kit, EZ2 PowerFecal Pro DNA/RNA Kit [5] [12] | Pre-packaged reagents and beads ensure consistency. Many include inhibitor removal technology, which is crucial for obtaining pure, amplifiable DNA from complex stool samples [5] [12]. |
| Sample Preservation | DNA/RNA Shield, PowerProtect DNA/RNA Reagent, RNAlater [5] [109] [12] | Stabilizes nucleic acids at room temperature immediately after collection, preventing degradation that could introduce pre-analytical variability and bias [5] [12]. |
| Homogenization | Bead-beating homogenizer (e.g., FastPrep-24) with lysing matrix tubes [5] | Mechanical lysis is essential for effective breakdown of diverse microbial cell walls (especially Gram-positive bacteria) in stool, ensuring representative and reproducible DNA yields [5]. |
| Quality Control | Qubit Fluorometer, NanoDrop Spectrophotometer [5] | Provides accurate quantification (Qubit) and purity assessment (NanoDrop) of extracted nucleic acids, which are critical data points for precision calculations [5]. |
| Reference Materials | ZymoBIOMICS Microbial Community Standard [5] | A defined mock community with known microbial composition. Serves as an ideal control material for assessing extraction efficiency, precision, and bias across multiple runs [5]. |
A meticulously executed reproducibility assessment is non-negotiable for establishing the reliability of automated nucleic acid extraction methods in stool sample research. By implementing the protocols outlined herein—adhering to CLSI guidelines, utilizing stable sample materials, incorporating multiple operators, and employing robust statistical analysis—researchers and drug development professionals can quantitatively demonstrate the inter- and intra-assay precision of their methods. The presented data and toolkit provide a framework for validating that automated systems deliver on their promise of high reproducibility, thereby ensuring that subsequent microbiome and pathogen data are of the highest quality and suitable for informing critical development decisions.
Conclusion
Automated nucleic acid extraction from stool samples has evolved from a technical challenge to an enabling technology for advanced molecular diagnostics and research. The integration of robust magnetic bead-based systems and emerging nanomaterials like Fe-MSN nanoparticles demonstrates significant improvements in yield, purity, and inhibition resistance. Successful implementation requires a thorough understanding of stool matrix complexities, optimized pre-analytical processing, and rigorous validation against both analytical and clinical standards. Future directions will likely focus on further miniaturization and integration of extraction with amplification and detection in fully automated 'sample-to-answer' systems, the development of even more resistant chemistries to overcome persistent inhibitors, and the creation of standardized protocols that ensure reproducibility across diverse clinical and research settings. As the field of microbiome-based diagnostics continues to mature, refined automated extraction methods will be fundamental to unlocking the full potential of stool as a rich, non-invasive source of molecular information for personalized medicine and public health.
References
- 1. New protocol for DNA extraction of stool [https://pubmed.ncbi.nlm.nih.gov/10683738/]
- 2. Methods for detection of Helicobacter pylori from stool ... sample [https://pmc.ncbi.nlm.nih.gov/articles/PMC8578210/]
- 3. US7005266B2 - Nucleic acid isolation from stool and other... samples [https://patents.google.com/patent/US7005266B2/en]
- 4. (PDF) New protocol for DNA extraction of stool [https://www.academia.edu/29207935/New_protocol_for_DNA_extraction_of_stool]
- 5. A Comparison of Three Automated Nucleic Acid Extraction Systems for Human Stool Samples - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11678849/]
- 6. of viral Extraction : comparison of five nucleic ... acids automated [https://pubmed.ncbi.nlm.nih.gov/22503856/]
- 7. Comparative evaluation of commercially available manual and automated nucleic acid extraction methods for rotavirus RNA detection in stools - PubMed [https://pubmed.ncbi.nlm.nih.gov/24036075/]
- 8. Quantitative Microbiome Profiling - Creative Biolabs [https://www.creative-biolabs.com/immuno-oncology/quantitative-microbiome-profiling.htm]
- 9. Isolation for Nucleic Acid Microbiome Research [https://www.thermofisher.com/us/en/home/life-science/dna-rna-purification-analysis/automated-purification-extraction/automated-magmax-kits-nucleic-acid-extraction/magmax-microbiome-extraction-kits.html]
- 10. Frontiers | A landscape review with novel criteria to evaluate microbial drivers for cancer: priorities for innovative research targeting excessive cancer mortality in sub-Saharan Africa [https://www.frontiersin.org/journals/cellular-and-infection-microbiology/articles/10.3389/fcimb.2025.1625818/full]
- 11. The tumor microbiome in cancer progression: mechanisms and therapeutic potential | Molecular Cancer | Full Text [https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-025-02403-w]
- 12. DNA or RNA isolation | Automation for stool samples | QIAGEN [https://www.qiagen.com/us/products/instruments-and-automation/automatable-kits/ez2-powerfecal-pro]
- 13. US4935342A - Method of isolating and purifying from... nucleic acids [https://patents.google.com/patent/US4935342A/en]
- 14. Nucleic Acid Extraction Techniques - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7122149/]
- 15. Next-Generation Sequencing Illumina Workflow–4 Key Steps | Thermo Fisher Scientific - US [https://www.thermofisher.com/us/en/home/life-science/cloning/cloning-learning-center/invitrogen-school-of-molecular-biology/next-generation-sequencing/illumina-workflow.html]
- 16. A rapid and high-yield method for nucleic acid extraction | Scientific Reports [https://www.nature.com/articles/s41598-025-95226-0]
- 17. Application of the NucliSENS easyMAG system for nucleic acid extraction: optimization of DNA extraction for molecular diagnosis of parasitic and fungal diseases - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3859032/]
- 18. 5 Best Practices for NGS Library Prep Success [https://dispendix.com/blog/5-best-practices-for-ngs-library-prep-success]
- 19. Isolation for Microbiome... | Thermo Fisher Scientific - ZA Nucleic Acid [https://www.thermofisher.com/za/en/home/life-science/dna-rna-purification-analysis/automated-purification-extraction/automated-magmax-kits-nucleic-acid-extraction/magmax-microbiome-extraction-kits.html]
- 20. A Comparison of Three Automated Nucleic Systems... Acid Extraction [https://www.mdpi.com/2076-2607/12/12/2417]
- 21. ... | Thermo Fisher Scientific - US Automated Nucleic Acid Extraction [https://www.thermofisher.com/us/en/home/life-science/dna-rna-purification-analysis/automated-purification-extraction/automated-magmax-kits-nucleic-acid-extraction.html]
- 22. - High -MGI Tech... throughput automated nucleic acid extractor [https://en.mgi-tech.com/products/instruments_info/17/]
- 23. Enhanced RNA Extraction from Stool Using Fe-MSN... Samples [https://brieflands.com/articles/apid-164719]
- 24. Next-generation Sequencing of the DNA Virome from Fecal Samples [https://bio-protocol.org/en/bpdetail?id=2159&type=0]
- 25. Selection of optimal extraction and RT-PCR protocols for stool RNA... [https://www.nature.com/articles/s41598-024-78680-0?error=cookies_not_supported&code=d5fad777-28fc-4a64-941f-b977d6884b12]
- 26. Assessment of nucleic for antibiotic... acid extraction protocols [https://humgenomics.biomedcentral.com/articles/10.1186/s40246-024-00617-5]
- 27. Silica-coated iron oxide nanoparticles for magnetic separation: polyol synthesis, superparamagnetic properties, and nucleic acid extraction efficiency | Journal of Nanoparticle Research [https://link.springer.com/article/10.1007/s11051-025-06467-z]
- 28. Impact of large-scale magnetic particle synthesis on nucleic acid extraction consistency and efficiency - ScienceDirect [https://www.sciencedirect.com/science/article/abs/pii/S0927776525004825]
- 29. Isolation of DNA from Arthrospira platensis and whole blood using... [https://jast-journal.springeropen.com/articles/10.1186/s40543-022-00337-2]
- 30. Comparative Evaluation of Different Surface Coatings of Fe 3O4-Based... [https://pubmed.ncbi.nlm.nih.gov/36012139/]
- 31. Comparison of commercial systems for extraction of nucleic ... acids [https://pmc.ncbi.nlm.nih.gov/articles/PMC7112907/]
- 32. Simplify Stool Sample Processing [https://www.alpco.com/resources/simplify-stool-sample-processing]
- 33. Beginners to Advanced Guide for Nucleic – Genetic... Acid Extraction [https://geneticeducation.co.in/beginners-to-advanced-guide-for-nucleic-acid-extraction/]
- 34. Rapid, single - tube method for quantitative preparation and analysis of... [https://bmcbiotechnol.biomedcentral.com/articles/10.1186/1472-6750-5-2]
- 35. A Single-Tube Nucleic Acid Extraction, Amplification, and Detection Method Using Aluminum Oxide - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1867575/]
- 36. A fully automatic and high-throughput centrifugal microsystem for rapid nucleic acid purification with a direct collection via detachable microtube interface - ScienceDirect [https://www.sciencedirect.com/science/article/abs/pii/S0925400525005234]
- 37. Comprehensive Stool Analysis by Mosaic Diagnostics (formerly Great...) [https://www.rupahealth.com/lab-tests/mosaic-diagnostics-comprehensive-stool-analysis]
- 38. Testing - The Ultimate In-Depth Guide (2021) - Healthpath Stool [https://healthpath.com/gut-health/stool-testing-the-ultimate-in-depth-guide/]
- 39. Market Automated and Forecast... Nucleic Acid Extraction Analysis [https://www.insightaceanalytic.com/report/automated-nucleic-acid-extraction-market/3281]
- 40. 2025 Automated Nucleic Acid Extraction Devices Industry Trends Report: Long-Term Outlook Through 2034 [https://www.openpr.com/news/4087715/2025-automated-nucleic-acid-extraction-devices-industry-trends]
- 41. Nucleic Acid Isolation and Purification Market Projected to Reach USD 16.38 Billion by 2032, Driven by Genetic Testing and Automation Trends, Reports SNS Insider [https://www.globenewswire.com/news-release/2025/07/14/3114831/0/en/Nucleic-Acid-Isolation-and-Purification-Market-Projected-to-Reach-USD-16-38-Billion-by-2032-Driven-by-Genetic-Testing-and-Automation-Trends-Reports-SNS-Insider.html]
- 42. Nucleic Acid Isolation and Purification Market Soars 9.56% CAGR by 2034 [https://www.towardshealthcare.com/insights/nucleic-acid-isolation-and-purification-market-sizing]
- 43. Devices Market Size Automated - 2034 Nucleic Acid Extraction 2025 [https://www.thebusinessresearchcompany.com/report/automated-nucleic-acid-extraction-devices-global-market-report]
- 44. China Customized Sterile Stool Collection... - CNWTC Sample [https://www.cnwtc.com/laboratory-plastic-disposables/specimen-container/sterile-stool-sample-collection-container.html]
- 45. The Simple - One for detection of... step stool processing method [https://pmc.ncbi.nlm.nih.gov/articles/PMC9531811/]
- 46. Optimization of the Simple One-Step Stool Processing Method to... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10434014/]
- 47. The Simple One-step stool processing method for... | PLOS One [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0264103]
- 48. Appendix . Xpert assays | MSF Medical Guidelines [https://medicalguidelines.msf.org/en/viewport/TUB/english/appendix-3-xpert-mtb-rif-20323734.html]
- 49. PCR inhibition in qPCR, dPCR and MPS—mechanisms and solutions - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7072044/]
- 50. Humic substances cause fluorescence inhibition in real-time polymerase chain reaction - PubMed [https://pubmed.ncbi.nlm.nih.gov/26170001/]
- 51. Characterization of the PCR effect of inhibitory to optimize... bile [https://pubmed.ncbi.nlm.nih.gov/15866213/]
- 52. as Complex in feces: Helicobacter... polysaccharides PCR inhibitors [https://pubmed.ncbi.nlm.nih.gov/9157172/]
- 53. (PDF) Complex as polysaccharides in feces... PCR inhibitors [https://www.academia.edu/16394324/Complex_polysaccharides_as_PCR_inhibitors_in_feces_Helicobacter_pylori_model]
- 54. A Comparison of Three Automated Nucleic Acid Extraction Systems for Human Stool Samples - PubMed [https://pubmed.ncbi.nlm.nih.gov/39770620/]
- 55. Evaluation of Inhibitor -Resistant Real-Time PCR for... Methods [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0073845]
- 56. & Enhancers – Dr. Pavan P. PCR Inhibitors [https://omegology.wordpress.com/2018/04/15/pcr-inhibitors-enhancers/]
- 57. Differing Effects of Standard and Harsh Nucleic ... Acid Extraction [https://pmc.ncbi.nlm.nih.gov/articles/PMC7916106/]
- 58. Processing faecal samples : a step forward for standards in microbial... [https://bmcmicrobiol.biomedcentral.com/articles/10.1186/1471-2180-14-112]
- 59. Recovery efficiencies of nucleic acid extraction kits as measured by quantitative LightCyclerTM PCR - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC1187008/]
- 60. CDC - DPDx - Diagnostic Procedures - Stool Specimens [https://www.cdc.gov/dpdx/diagnosticprocedures/stool/specimenproc.html]
- 61. Comparison of Nucleic Acid Extraction Methods for a Viral Metagenomics Analysis of Respiratory Viruses [https://www.mdpi.com/2076-2607/8/10/1539]
- 62. Transforming Nucleic with Acid | The Scientist Extraction Automation [https://www.the-scientist.com/transforming-nucleic-acid-extraction-with-automation-72801]
- 63. DNA and RNA Automating - Analytik Jena Extraction [https://www.analytik-jena.us/knowledge/topics/molecular-biology/automated-dna-isolation-choosing-a-method/]
- 64. of Optimization Buffer Composition (Polyethylene Glycol, Sodiu Binding [https://www.dovepress.com/optimization-of-binding-buffer-composition-polyethylene-glycol-sodium--peer-reviewed-fulltext-article-NSA]
- 65. Optimizing Stringency in Wash Buffers for Hybridization Assays - CSIR NET LIFE SCIENCE COACHING | NTA NET LIFE SCIENCE | CSIR LIFE SCIENCE [https://www.letstalkacademy.com/increasing-stringency-hybridization-buffers/]
- 66. ELISA Plate Washing Optimization | Boster Bio [https://www.bosterbio.com/protocol-and-troubleshooting/picokine-elisa-optimization/washing]
- 67. quantification and RNA assessment techniques | QIAGEN quality [https://www.qiagen.com/us/knowledge-and-support/knowledge-hub/bench-guide/rna/rna-analysis/quantification-and-analysis-of-rna]
- 68. A Guide to DNA and RNA Quantification and... | Technology Networks [https://www.technologynetworks.com/genomics/articles/a-guide-to-dna-and-rna-quantification-and-quality-392900]
- 69. Methods of RNA Assessment Quality [https://pl.promega.com/resources/pubhub/methods-of-rna-quality-assessment/]
- 70. Validating RNA Quantity and Quality: Analysis of RNA Yield, Integrity, and Purity | BioPharm International [https://www.biopharminternational.com/view/validating-rna-quantity-and-quality-analysis-rna-yield-integrity-and-purity]
- 71. to Check Methods | Thermo Fisher Scientific - SG RNA Integrity [https://www.thermofisher.com/sg/en/home/references/ambion-tech-support/rna-isolation/tech-notes/is-your-rna-intact.html]
- 72. integrity and the effect on the real-time qRT-PCR performance RNA [https://pubmed.ncbi.nlm.nih.gov/16469371/]
- 73. RNA analysis by ion-pair reversed-phase high performance liquid chromatography - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC29688/]
- 74. Bleach Gel: A Simple Agarose Gel for Analyzing RNA Quality - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3699176/]
- 75. Optimizing 16S rRNA gene profile analysis from low biomass nasopharyngeal and induced sputum specimens | BMC Microbiology | Full Text [https://bmcmicrobiol.biomedcentral.com/articles/10.1186/s12866-020-01795-7]
- 76. Organic Extraction Method | Thermo Fisher Scientific - US [https://www.thermofisher.com/us/en/home/life-science/dna-rna-purification-analysis/organic-extraction.html]
- 77. , Part One - Westgard QC Probit Analysis [https://westgard.com/lessons/basic-method-validation/probit-part-one.html]
- 78. GitHub - sahoomk/LLoD_ Probit : Calculate Lower limit ... of detection [https://github.com/sahoomk/LLoD_Probit]
- 79. Cytomegalovirus DNA Quantification Using an Automated Platform for Nucleic Acid Extraction and Real-Time PCR Assay Setup - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3147859/]
- 80. Extraction of viral nucleic acids: Comparison of five automated nucleic acid extraction platforms - ScienceDirect [https://www.sciencedirect.com/science/article/abs/pii/S1386653212001254]
- 81. Comparative Assessment of Automated Nucleic Acid Sample Extraction Equipment for Biothreat Agents - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3993465/]
- 82. of Sensitivity estimation using analysis ... limit of detection probit [https://pubmed.ncbi.nlm.nih.gov/41240387/]
- 83. Automated nucleic acids purification from fecal samples on a microfluidic cartridge | BioChip Journal [https://link.springer.com/article/10.1007/s13206-016-1205-5]
- 84. Use and Interpretation of Enteropathogen Multiplex Nucleic Acid Amplification Tests in Patients With Suspected Infectious Diarrhea - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6284344/]
- 85. Frontiers | A Wipe-Based Stool Collection and Preservation Kit ... [https://frontiersin.org/articles/10.3389/fimmu.2022.889702/full]
- 86. yeast promotes Salmonella Typhimurium virulence | Nature Commensal [https://www.nature.com/articles/s41586-025-09415-y?error=cookies_not_supported&code=0ec8d043-e3c1-4397-894c-ad6c0f4d44a1]
- 87. Ocular pathogen or commensal : a PCR-based study of surface... [https://pubmed.ncbi.nlm.nih.gov/18055811/]
- 88. Automated microfluidic system for rapid, multiplexed nucleic acid extraction and detection of pathogens - ScienceDirect [https://www.sciencedirect.com/science/article/abs/pii/S0925400524017386]
- 89. Comparison of Automated and Manual Nucleic Acid Extraction Methods for Detection of Enterovirus RNA - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC179781/]
- 90. Current Nucleic Acid Extraction Methods and Their Implications to Point-of-Care Diagnostics - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC5529626/]
- 91. Purification Methods: Phenol-Chloroform... Comparing Nucleic Acid [https://www.lunanano.ca/post/comparing-nucleic-acid-purification-methods-phenol-chloroform-silica-spin-column-magnetic-beads]
- 92. Bio-On-Magnetic-Beads (BOMB): Open platform for high-throughput nucleic acid extraction and manipulation - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6343928/]
- 93. Method: The Best Detection Method for Pathogens... Magnetic Bead [https://www.lingjunbio.com/pathogen-detection-using-magnetic-beads/]
- 94. A rapid and high-yield method for nucleic | Scientific... acid extraction [https://www.nature.com/articles/s41598-025-95226-0?error=cookies_not_supported&code=e9fc88e8-b0f9-48ba-9f79-0654dc23ea1c]
- 95. Rapid Equipment-Free Nucleic Using a Acid -Based... Extraction Silica [https://pmc.ncbi.nlm.nih.gov/articles/PMC12120609/]
- 96. Automated magnetic technology bead [https://www.news-medical.net/whitepaper/20250424/Automated-magnetic-bead-technology-for-high-throughput-viral-nucleic-acid-isolation.aspx]
- 97. What is Magnetic -based Bead ? Uses, How It... Nucleic Acid Extraction [https://www.linkedin.com/pulse/what-magnetic-bead-based-nucleic-acid-extraction-ig2lc]
- 98. Magnetic particles for integrated nucleic acid purification, amplification and detection without pipetting - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7202819/]
- 99. Isolation: Strategies for Reliable Sequencing... Optimizing Nucleic Acid [https://www.linkedin.com/pulse/optimizing-nucleic-acid-isolation-strategies-reliable-sequencing-fbmye]
- 100. Comparison of three rapid and easy bacterial DNA extraction methods for use with quantitative real-time PCR - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3181010/]
- 101. Frontiers | Comprehensive performance evaluation of... [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1609142/full]
- 102. evaluation of the Performance NucliSens easyMAG... automated [https://pubmed.ncbi.nlm.nih.gov/19500527/]
- 103. Development and validation of a rapid five-minute nucleic acid extraction method for respiratory viruses | Virology Journal | Full Text [https://virologyj.biomedcentral.com/articles/10.1186/s12985-024-02381-3]
- 104. Evaluation of automatic cell free DNA extraction metrics using different blood collection tubes | Scientific Reports [https://www.nature.com/articles/s41598-025-03508-4]
- 105. Evaluating Assay - PMC Precision [https://pmc.ncbi.nlm.nih.gov/articles/PMC2556577/]
- 106. 4.1. Repeatability, intermediate precision and reproducibility – Validation of liquid chromatography mass spectrometry (LC-MS) methods [https://sisu.ut.ee/lcms_method_validation/41-precision-trueness-accuracy/]
- 107. Calculating Inter- and Intra-Assay Coefficients of Variability – Salimetrics [https://salimetrics.com/calculating-inter-and-intra-assay-coefficients-of-variability/]
- 108. Instruments - Amerigo Scientific Automated Nucleic Acid Extraction [https://www.amerigoscientific.com/instrument/automated-nucleic-acid-extraction-instruments.html]
- 109. total Automated with magnetic beads for the... nucleic acid extraction [https://pmc.ncbi.nlm.nih.gov/articles/PMC11668005/]